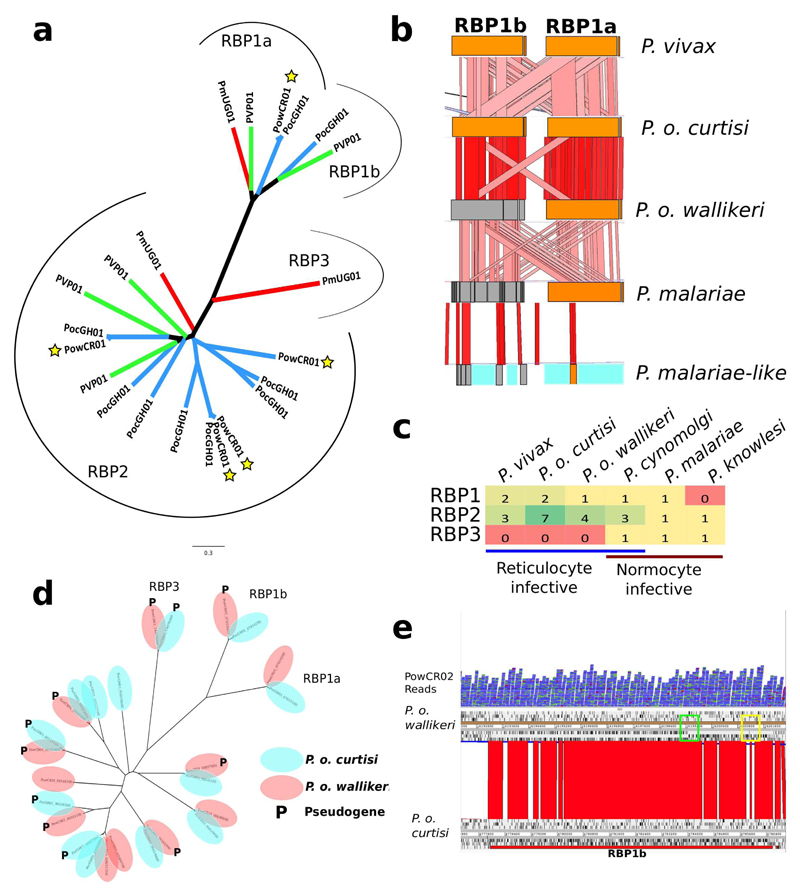
Extended Data Figure 4. Reticulocyte-binding protein changes in P. malariae and P. ovale.
a, Phylogenetic tree of all full-length functional RBPs in P. malariae (red branches), P. o. curtisi (blue branches without stars), P. o. wallikeri (blue branches with stars), and P. vivax (green branches). Brackets indicate the different subclasses of RBPs: RBP1a, RBP1b, RBP2 and RBP3. b, ACT41 view of functional (orange) and pseudogenized (grey) RBP1a and RBP1b in five species (P. vivax, P. o. curtisi, P. o. wallikeri, P. malariae and P. malariae-like). Blue indicates assembly gaps. Red bars between species indicate level of sequence similarity, with darker colour indicating higher similarity. c, Number of RBP genes in each of the three RBP classes (RBP1, RBP2, RBP3) by species (P. vivax, P. o. curtisi, P. o. wallikeri, P. cynomolgi, P. malariae, P. knowlesi) grouped by erythrocyte invasion preference (reticulocyte versus normocyte). d, PhyML50 generated phylogenetic tree of all RBP genes over 1 kb long in P. o. curtisi and P. o. wallikeri. Pseudogenes are denoted with (P). Multiple functional RBP2 genes match up with pseudogenized copies in the other genome. e, ACT41 view of RBP1b in red for P. o. curtisi (bottom) and the corresponding disrupted open reading frame in P. o. wallikeri (top), with black ticks indicating stop codons. Reads (in blue) from an additional P. o. wallikeri sample (PowCR02) confirm the bases introducing the frameshift (green box) and premature stop codon (yellow box) in RBP1b.