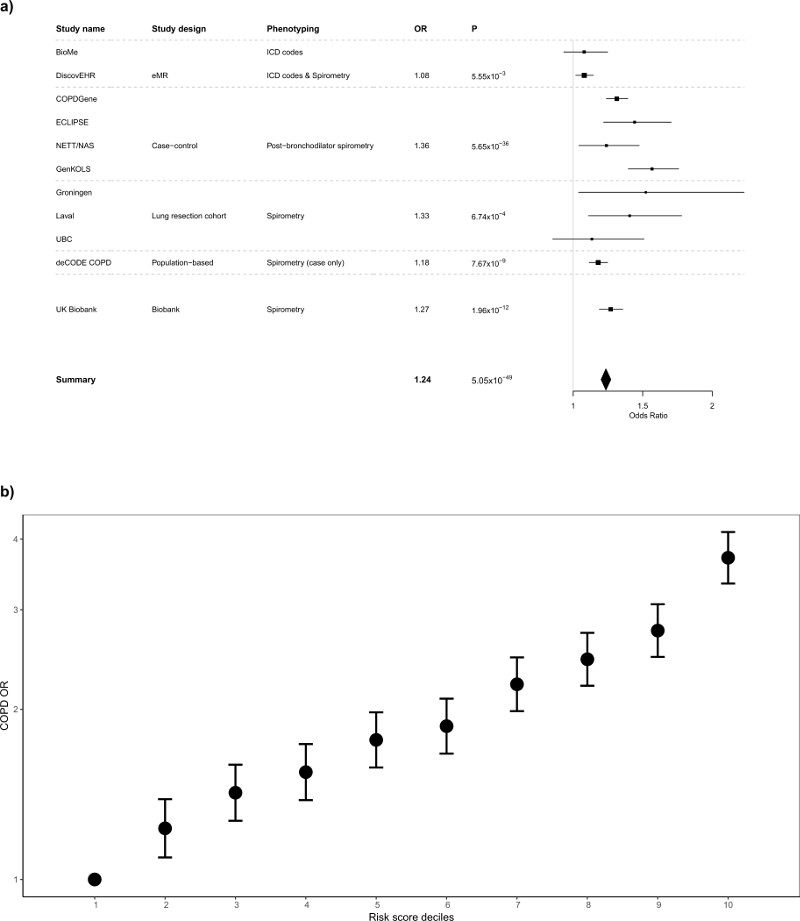
Figure 2.
Genetic Risk Score associations with COPD susceptibility (a) Forest plot of COPD results for the risk score analysis. Odds ratios per standard deviation of the risk score (~6 alleles) are presented for each study. Studies are grouped according to study design and phenotyping: “eMR”, electronic medical records, which used ICD codes to define COPD (DiscovEHR also used spirometry to refine the COPD definition); “case-control”, COPD case-control, which used post-bronchodilator spirometry to define COPD; “lung resection cohort”, which used a combination of pre and post-bronchodilator spirometry to define COPD; the Icelandic Biobank, deCODE, where cases were selected from a population based study and a study of COPD patients and defined using a spirometric definition, controls were selected as individuals within the cohort that were not known cases (no spirometric definition was used for controls); and UK Biobank (excluding UK BiLEVE), which used spirometry to define both COPD cases and controls. Further details are provided in the Supplementary Note. (b) Odds ratios for spirometrically-defined COPD for weighted genetic risk score deciles in UK Biobank (10,547 cases, pre-bronchodilator % predicted FEV1<80% and FEV1/FVC<0.7, and 53,948 controls, FEV1/FVC>0.7 and % predicted FEV1>80%, weights derived from non-discovery populations). For each decile, odds ratios were obtained using a logistic regression adjusted for age, age2, sex, height, smoking status, pack-years and the first 10 ancestry principal components. The OR comparing the 10th and the 1st decile in ever-smokers only was 3.35 (95% CI 2.93 to 3.84) and in never-smokers only was 4.27 (95% CI 3.61 to 5.06).