Figure 1.
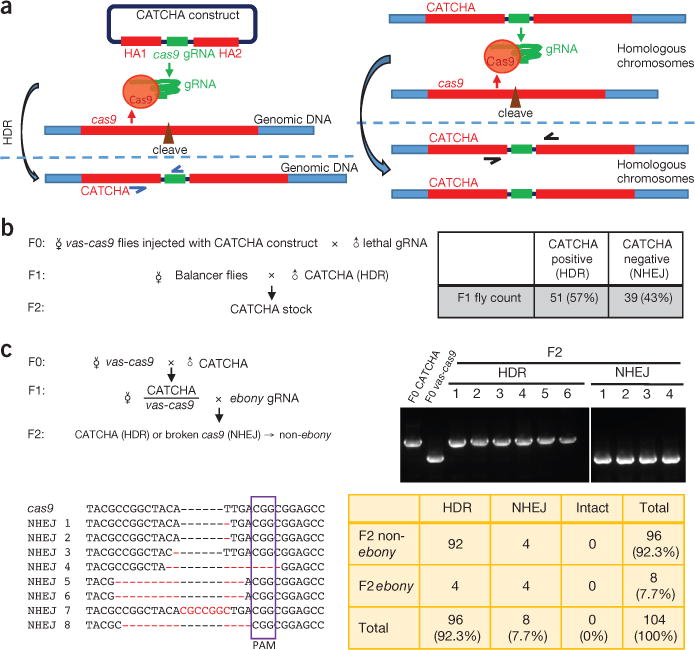
Schematics and proof of principle of CATCHA. (a) Left, the CATCHA construct encodes a gRNA targeting cas9, flanked by homology arms (HAI and HA2) corresponding to cas9 sequences next to the cleavage site specified by the gRNA (see Supplementary Methods for details). In the presence of genomically encoded Cas9, the CATCHA construct is integrated into the target locus via HDR. Successful HDR events can be identified by PCR with suitable primers (blue). Right, the genomically integrated CATCHA can further convert allelic cas9 sequences to CATCHA via HDR. Indels and converted alleles can be distinguished with PCR primers (black arrows). Primer information is available in Supplementary Methods. (b) Integrating CATCHA into the genome. vas-cas9 donor females injected with the CATCHA construct were crossed to males expressing gRNA targeting wg to select for ablated cas9 alleles. PCR was performed for each F1 survivor to identify CATCHA-positive progenies and establish stable stocks. NHEJ is presumed on the basis of PCR results. Balancers are fly chromosomes with multiple inversions that impede recombination and thereby maintain transgenes and mutations in stable stocks. (c) Converting genomic cas9 using genomic CATCHA. All 104 F2 offspring from one CATCHA male crossed to vas-cas9 females were genotyped using PCR. Representative DNA gel lanes are shown (top right). F0 CATCHA and F0 vas-cas9 are controls indicating the respective sizes of CATCHA (~1,200 bp) and NHEJ (~650 bp) bands. In sum, 96 F2 flies carried CATCHA, suggesting a conversion rate of 85%, estimated by the formula (CATCHA - total × 50%) / (total × 50%). All eight F2 flies with short bands (non-HDR) were sequenced (bottom left) and confirmed to carry NHEJ-mediated indels (nucleotides differing from cas9 sequence are shown in red). The four indels that maintain the reading frame (NHEJ 5–8) correspond to the four ebony flies. The purple box highlights the protospacer adjacent motif (PAM) sequence. See Supplementary Table 1 for the summary of all F2 phenotypes.
