Figure 4.
Validation of the A-to-I Editing Model and Quantitative Estimation of Site-Specific Editability
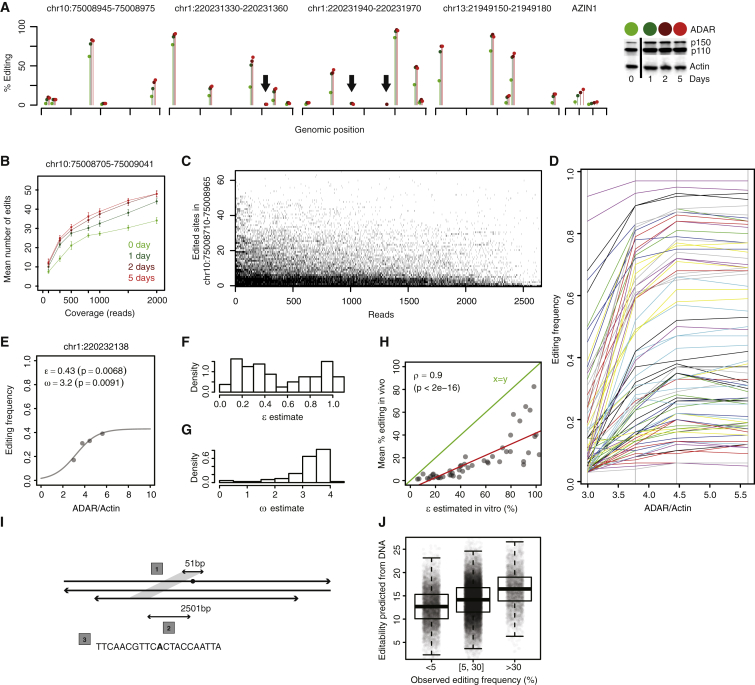
(A) Effect of increasing ADAR expression in the cell line MCF7 on editing in four representative Alu regions and AZIN1. The full-length of sequenced regions are shown in Figure S3 for MCF7 and three more cell lines. Complete ADAR western blots quantifications underlying the color scale are provided in Figure 5E (see baseline t = 0 and IFN-α, t ∈ {1, 2, 5} days tracks) and in Figure S7. Increasing ADAR expression increases the editing frequency at all editable positions, as predicted by the model of Figure 3D. Similar results were obtained for IFN-β and IFN-γ (global, position-less, view Figure 5F). Arrows point at editing sites detectable only at higher ADAR expression in our assay.
(B) Increasing sequencing coverage (x axis) or ADAR expression (color scale) increases the number of detectable editing sites (y axis). Coverage variation was implemented by down-sampling the total pool of sequencing reads, starting from 2,000×, down to 100×, and re-running the variant detection pipeline for each down-sampled alignment. Each data point is the mean of 30 down-sampling experiments. Error bars, SD.
(C) Editing of individual mRNA molecules. Each black dot depicts an edited base in a given mRNA molecule. The y axis goes from 0 to 60 and corresponds to the adenosines in the ∼250-bp span that are edited in at least one of the 2,842 reads represented along on the x axis. Reads and adenosines were ordered by decreasing editing frequencies. 185 non-edited reads were omitted from the figure.
(D) Dose-response curves for experiment in cell line BT474. ADAR was increased through IFN-α stimulation (as in A). We focused on 81 sites (color lines) with a baseline editing frequency >2.5% in order to avoid trivial nonlinear effects caused by lack of detection at low ADAR expression.
(E) Example of a fit of the logistic model (line) to experimental data points (dots). The unit of ω is commensurate to the dimensionless ADAR relative expression and ε is the fraction of edited transcripts at saturation.
(F and G) Distributions of ε and ω across the 81 sites.
(H) The 81 edited sites are depicted as dots with the corresponding εi estimates derived from the BT474 cell lines on the x axis and their in vivo editing frequency on the y axis.
(D)–(H) are part of a more comprehensive analysis presented in Figure S4.
(I) DNA-based statistical model of editability. The model included three parameters: (1) the best Smith-Waterman global alignment score of the 51-bp sequence surrounding the editing site (green dot) within the 2,501-bp sequence surrounding the editing site on the reverse strand; (2) the distance separating the editing site from this best alignment; (3) the 20 nucleotides surrounding the editing site. These 1 + 1 + 20 = 22 variables were fitted with a linear model against the editing frequencies of half of 51,621 Alu editing sites with coverage ≥20× previously identified (Ramaswami et al., 2012).
(J) Observed editing frequencies versus editabilities predicted from DNA for validation sites.