Figure 5.
ADAR Amplification and the Interferon Response Are Independent Predictors of ADAR Expression in Cancer
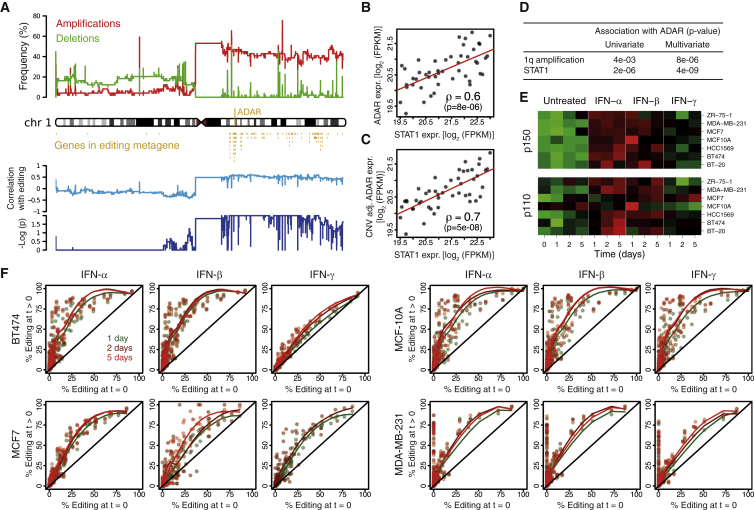
(A) The top panel shows the frequencies of amplifications/deletions along chromosome 1 in our series. The middle panel shows the genes whose expression is highly associated with that of ADAR. Nineteen genes not located on chr1 are omitted. The bottom panel shows the Spearman’s correlation coefficient and associated p values of non-segmented copy-number array probes with the sample-wise mean editing frequencies.
(B) Dots represent tumor samples, with STAT1 expression on the x axis and ADAR expression on the y axis.
(C) Same as (B) with ADAR expression adjusted for ADAR copy number.
(D) Association p values of ADAR copy number and STAT1 expression with ADAR expression increase in a multivariate analysis, demonstrating that ADAR expression is independently associated with these two variables.
(E) Seven breast cancer cell lines were exposed to interferon α, β, and γ for 1, 2, and 5 days. Western blots quantifications are depicted for each cell line, interferon, and time. Because expression dynamic ranges vary among cell lines, each line has its own color scale extending from low expression in green to high expression in red. The underlying gels are presented in Figure S7 and blot quantification in Table S6. Corresponding mRNA RT-PCR expression data are shown Figure S7 and detailed Table S7.
(F) Editing frequencies in the absence of treatment (x axis) versus interferon treatment (y axis). Points depict the editing sites in AZIN1 and the four Alu regions of Figure S3. Points are above the identity line x = y (black diagonals); i.e., interferons increase editing frequencies at all sites. Library preparation failed for MCF7/IFN-γ at 5 days. Limited sequencing coverage precluded detection of some editing events for MDA-MB-231, t = 0 and t = 1 days.