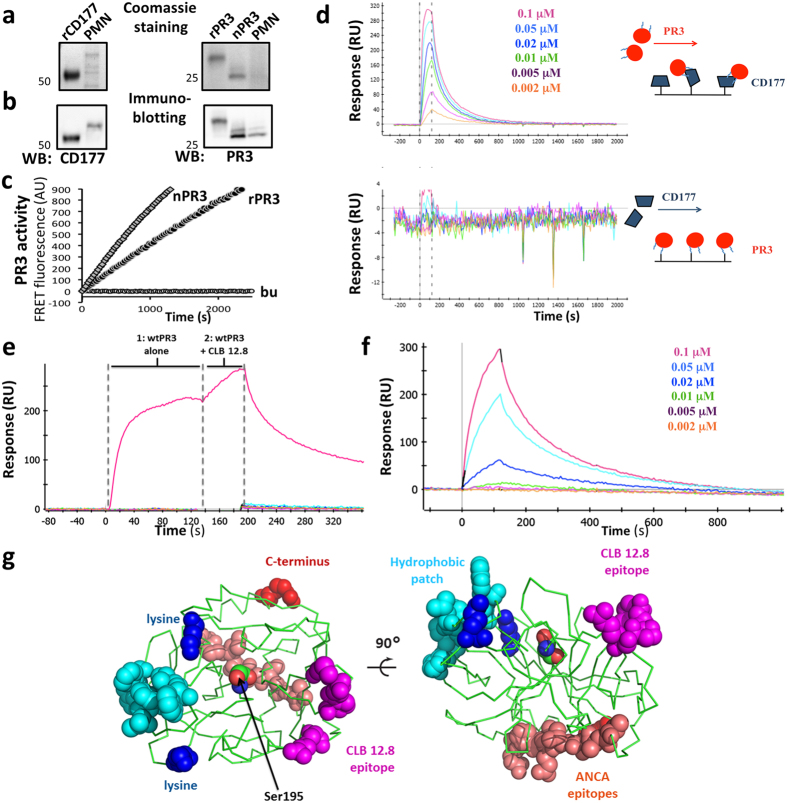
Figure 3. Characterization of rCD177, rPR3 and nPR3, and of the PR3:CD177 interaction.
rCD177, rPR3, nPR3, and neutrophil lysates (PMN) were subjected to non-reducing SDS PAGE, (a) Coomassie stained and (b) immunoblotted. rCD177 lacks the C-terminal GPI-anchor domain and therefore migrates faster than nCD177. rPR3 has 12 additional (assumed unstructured) C-terminal residues that affect migration on the SDS gel compared to nPR3. The molecular weights are indicated by the numbers to the left of each image. (c) Enzymatic activity of 1 nM PR3 was measured by FRET assay in the presence of 0.02% lauryl maltoside (LM). (d) SPR sensorgrams showing the PR3:CD177 interaction. Vertical dotted lines indicate ligand flow across the chip. Top, CD177 was immobilized to the sensor chip and PR3 at the indicated concentrations was the soluble analyte; high affinity complex formation is observed. Bottom, the same experiment with the reverse configuration: no interaction is observed. Blue lines on the PR3 schematic represent the two lysines available for PR3 immobilization. PR3 is almost certainly uniformly immobilized with its active site facing the chip surface. (e,f) Restricting the position of the PR3:CD177 interface. (e) SPR sensorgram of the three-way interaction between PR3, CLB 12.8 and CD177. CD177 was immobilized on the sensor chip and subjected to 2 back-to-back ligand flows (horizontal dotted lines). First, soluble PR3 alone was allowed to flow for 120 seconds, producing a resonance units (RUs) increase as PR3 bound CD177; next, a mixture of PR3 and CLB 12.8 was allowed to flow over the chip for 60 seconds and showed an additional RU increase, indicating that CLB 12.8 can bind the complex and, therefore, that the CD177 and CLB 12.8 epitopes do not overlap. (f), Sensorgram of the monomeric PR3 hydrophobic patch mutant (Ile217Ala, Trp218Ala) binding to immobilized CD177. (g) Orthogonal views of PR328 showing key surface features (rendered using the PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC). The backbone is shown in ribbon representation, the hydrophobic patch residues in cyan, the two lysines (Lys) used for immobilization in SPR in blue, the CLB 12.8 epitope residues in magenta, the C-terminal Arg in red and a stretch containing some of the most common human ANCA epitopes38 in salmon. The catalytic serine 195 is also shown as spheres.