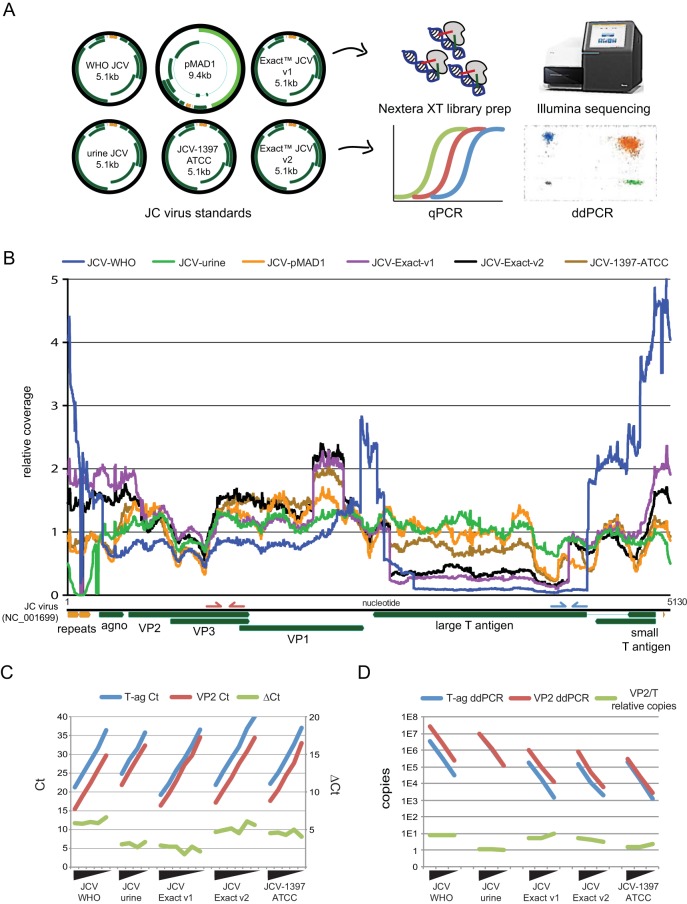
FIG 1.
Next-generation sequencing of JC virus standards reveals deletions and copy number heterogeneity. (A) Six different JC virus materials were deep sequenced, and five standards were tested by qPCR and ddPCR. Gene organization of each JC virus is shown in green, with regulatory regions depicted in orange. Of note, the 9.4-kb pMAD1 plasmid inserted the backbone at the location of the probe used in the T antigen qPCR and ddPCR assay and cannot be quantitated at that locus. (B) Coverage plot of six different standards of JC virus mapped to the JC virus NCBI reference genome (GenBank accession number NC_001699). The y axis is normalized such that 1 designates the average coverage across the viral genome to highlight relative differences in coverage. Three of the six standards include a large deletion in the T antigen region that constitutes a greater than 4-fold difference in copy number relative to the structural genes. Reductions in coverage in the regulatory repeat region are due both to small deletions and sequence divergence relative to JC virus reference genome. Primers for the Focus PCR analyte-specific reagent targeting the VP2/3 region are shown on the JC virus genome in red, while the pep primers targeting the T antigen region are shown in blue. (C, D) Confirmation of the copy number differences seen by sequencing was performed with qPCR (C) and ddPCR (D) using PCR primers against the VP2/3 gene (red) and T-ag gene (blue). Ten-fold dilutions of each of the standards depicted were quantitated, and the discrepancy in cycle threshold (CT) and absolute quantitation (D) between the VP2/3 and T-ag assays are depicted (green) for each standard. ΔCT, change in CT.