FIG 1.
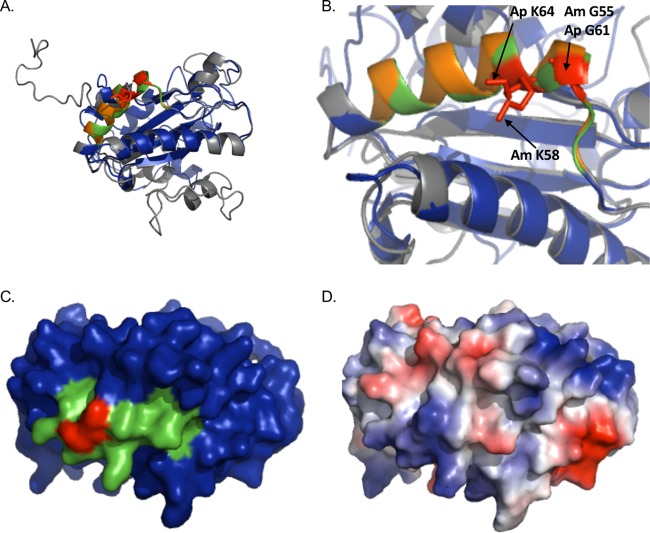
AmOmpA and ApOmpA are structurally similar and exhibit conservation of glycine and lysine residues demonstrated to be important for adhesin function in ApOmpA. (A and B) The predicted tertiary structures for ApOmpA and AmOmpA are highly similar. (A) Presented is a static image from Movie S1 in which the predicted tertiary structures for ApOmpA (gray) and AmOmpA (blue) are overlaid to demonstrate their structural similarity. A PHYRE2 model of the mature sequence lacking signal peptide for each OmpA protein was generated, and the models were threaded onto each other using PyMOL. ApOmpA residues 59 to 74, which comprise the essential binding domain, are orange. AmOmpA residues 53 to 68, which form an alpha-helix that is similar in location to that formed by ApOmpA essential binding domain residues 59 to 74 and are therefore predicted to form the AmOmpA binding domain, are green. ApOmpA residues glycine 61 and lysine 64, which were previously confirmed to be essential for adhesin function, are red. (B) Zoom-in of the image presented in panel A. Note that the alpha helices formed by the essential binding domain of ApOmpA and the putative AmOmpA binding domain overlap. ApOmpA functionally essential residues glycine 61 and lysine 64 correspond to AmOmpA G55 and K58. (C) Space-filling model of AmOmpA. The putative binding domain encompassed by residues 53 to 68 is indicated in green and G55 and K58 are red. (D) Electrostatic surface map of A. marginale OmpA, as generated using the PyMol APBS plugin. Positive and negative charges are indicated by blue and red, respectively.