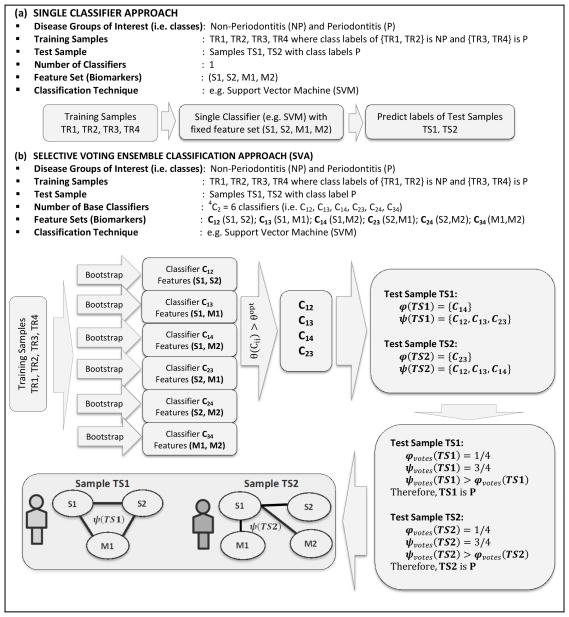
Figure 1.
Working principle of traditional single classifier system C using salivary (S1, S2) and microbial (M1, M2) biomarkers simultaneously as features in the classification process is shown in (a) whereas those of SVA using pairs of biomarkers as features and an ensemble of classifiers {C12,C13,C14,C23,C24,C34} is shown in (b). Imposing the optimal sensitivity threshold in SVA returns a subset of classifiers {C12,C13,C14,C23}. The training samples (TR1,TR2,TR3,TR4) were used to learn the classifiers and used subsequently to predict the labels of the test samples (TS1,TS2). The normalized vote-counts of the periodontitis test samples TS1 and TS2 using majority voting is identical , indicating equal proclivity of TS1 and TS2 to periodontitis group. However, network abstractions of their ensemble sets exhibit markedly different topologies revealing patient-specific variations within the periodontitis subjects and cross-talk between the salivary and microbial biomarkers.