Figure 2.
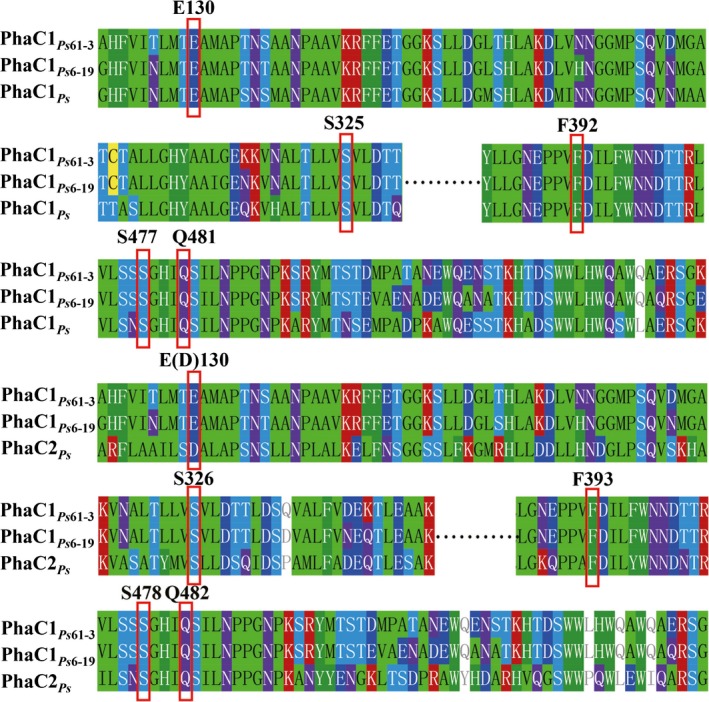
Alignment of phaC1 Ps and phaC2 Ps with previous LA polymerizing enzymes reported (Taguchi et al., 2008; Yamada et al., 2010; Yang et al., 2011). phaC1 Ps and phaC2 Ps from Pseudomonas stutzeri were aligned with reported LA polymerizing enzymes which were phaC1 Ps61‐3 from Pseudomonas sp. 61‐3 and phaC1 Ps6‐19 from Pseudomonas sp. MEBL6‐19. Several constitutive sites were found dominating the LA polymerizing capacity (in the red frames). Abbreviations: phaC1Ps, Pseudomonas stutzeri phaC1; phaC2Ps, Pseudomonas stutzeri phaC2; phaC1Ps61‐3, Pseudomonas sp. 61‐3 phaC1; phaC1Ps6‐19, Pseudomonas sp. MEBL6‐19 phaC1; E, glutamic acid; D, aspartic acid; S, serine; F, phenylalanine; Q, glutamine. Five mutation sites on phaC1 ps (phaC2 ps), including E130D, S325(326)T, F392(393)S, S477(478)G and Q481(482)K, were selected based on the alignment result with the LA polymerizing enzymes.
