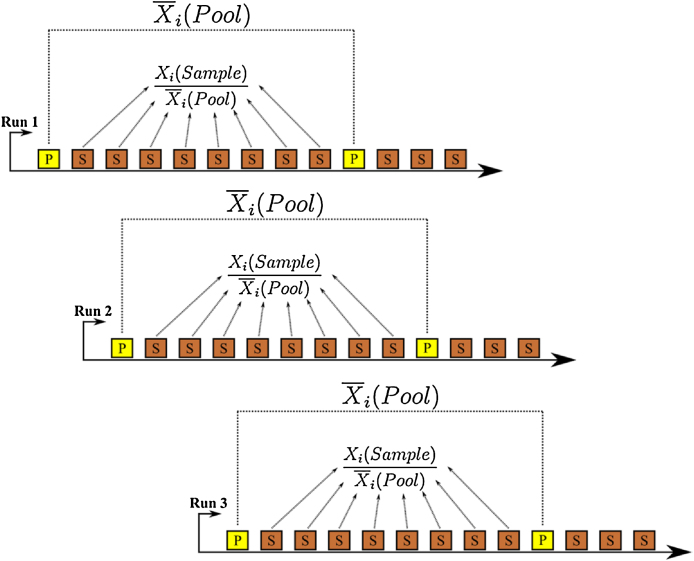
Fig. 1.
Normalization by reference pool measurements. In a GC–MS sequence plan, every 8th position a reference pool sample is measured. For pool normalization, the average of the 2 nearest pools is calculated for each metabolite i (). Then, for each sample the metabolite intensity of each metabolite i is divided by the corresponding average of the pools: .