Fig. 1.
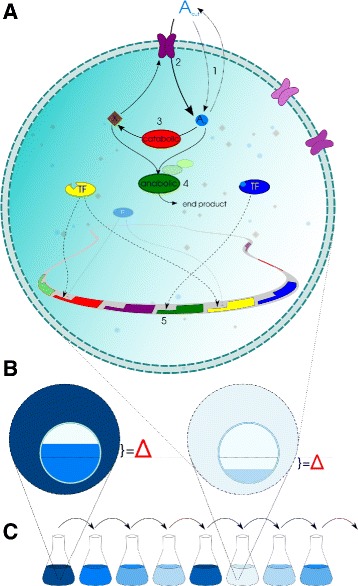
Virtual Cell model overview. a Virtual Cells have a circular genome that encodes metabolic and regulatory proteins. An externally available resource molecule (A) diffuses passively over the membrane (1) and is actively imported (2) by pump proteins. Once inside, A is converted to (X) by catabolic enzymes. X serves as the energy source for the import reaction (2). In addition, A and X are converted to an unspecified end product (4) by anabolic enzymes. Protein expression from genes (5) can be regulated by TFs if their binding motif matches the gene’s operator sequence. Binding of a ligand (A or X) by the TF alters its regulatory effect on gene expression. The genome can contain multiple copies of any of the gene types. Different copies may encode different values of the gene’s parameters, such as the enzymatic constants of the reaction that they catalyse or the binding motif and regulatory effect. b Fitness is determined by measuring the difference (Δ) between the realised steady state concentrations of internal A and X and the homeostasis target value (dotted line). c During the evolutionary experiments the external concentration of A is continually varying, while the homeostasis target remains constant. Cells have a chance proportional to their fitness to contribute offspring to the next generation
