Abstract
It has been appreciated for decades that somatic genomic alterations that change coding sequences of proto-oncogenes, translocate enhancers/promoters near proto-oncogenes, or create fusion oncogenes can drive cancer by inducing oncogenic activities. An explosion of genome-wide technologies over the past decade has fueled discoveries of the roles of three-dimensional chromosome structure and powerful cis-acting elements (super-enhancers) in regulating gene transcription. In recent years, studies of human T cell acute lymphoblastic leukemia (T-ALL) using genome-wide technologies have provided paradigms for how non-coding genomic region alterations can disrupt 3D chromosome architecture or establish super-enhancers to activate oncogenic transcription of proto-oncogenes. These studies raise important issues to consider with the objective of leveraging basic knowledge into new diagnostic and therapeutic opportunities for cancer patients.
Non-Coding Genomic Sequences Regulate Gene Transcription
The intrinsic composite expression of genes defines cellular identity, function, and growth. For decades, it has been known that inherited and somatic genomic alterations that change coding sequences of proto-oncogenes (see Glossary), translocate heterologous promoters/enhancers near proto-oncogenes, or generate fusion oncogenes cause malignant cellular transformation by unleashing oncogenic activities. It has also been recognized that elevated expression of proto-oncogenes in the absence of detectable alterations at these loci is common in human cancer cells, implying other undetermined mechanisms for oncogene activation. Instances of aberrant monoallelic transcription of proto-oncogenes in cancer cells had suggested a role of cis-acting factors in promoting malignant transformation. However, until recently, the inability to interrogate entire genomes and epigenomes had been a barrier to identifying additional pathological mechanisms activating oncogenic transcription of proto-oncogene loci.
An explosion of next generation sequencing (NGS)-based genome-wide techniques (Table 1) during the last decade smashed this barrier and provided unprecedented insights into how genomic alterations of non-coding regions inappropriately activate gene transcription. Comparisons among whole genome sequences of humans afflicted with various diseases with healthy human controls revealed that many inherited disease-linked genomic variants, including some associated with cancer predisposition, actually reside in non-coding regions [1–6]. Most of these nucleotide differences occur in non-coding regions defined as enhancers based on their overlaying chromatin profiles identified by ChIP-Seq (Table 1)[7], which combines NGS with chromatin immunopreciptiation (ChIP) [8] to determine genomic sequences where a protein or protein modification resides (Table 1). The Hi-C method (Table 1) [9], which couples NGS with chromosome conformation capture (3C) assays [10] (Table 1), has been used to identify all pairwise genomic interactions (Table 1), showing that mammalian genomes partition into megabase (Mb)-sized topological association domains (TADs) in which sequences interact more frequently with one another than with sequences in other TADs [11]. TADs are conserved among species, cell types, and cell developmental stages, with TAD borders often enriched for binding of the CCCTC-binding factor (CTCF) chromosome structural protein [11, 12]. Additional experiments have shown that TADs subdivide into structures composed of chromosome loops formed by interactions between CTCF binding elements (structural loops) or between promoters and enhancers (regulatory loops), with both types of loops stabilized by binding to cohesin proteins [13]. Such intra-TAD structures vary among cell types and stages, correlating with the expression of their composite genes [14]. Structural loops often compartmentalize enhancers and their genes to insulate them from communicating with promoters and enhancers, respectively, in adjacent structural loops [15, 16]. Disease-associated genomic variants have been found to disrupt CTCF binding sites, breaking architectural borders and allowing inappropriate functional interactions between enhancers and promoters, normally insulated within separate domains [17–19]. Furthermore, ChIP-Seq has revealed that some non-coding regions of mammalian genomes exhibit -- in a cell type-specific manner -- abnormally high levels of bound transcription factors and of histone modifications associated with active transcription [20, 21]. In addition, the distances over which these factors bind and histone modifications occur are much larger than typical enhancers [20, 21]. These non-coding genomic regions have been termed super-enhancers (or stretch-enhancers), which orchestrate high-level gene transcription by activating target promoters [20].
Table 1.
NGS-based Genome-Wide Technologies
| Technique | Brief Description | Result | Refs. |
|---|---|---|---|
| ChIP: Chromatin Immunoprecipitation | Cells are treated to cross-link DNA with associated proteins. DNA-protein complexes are fragmented by sonication or nuclease digestion and subject to immunoprecipitation with an antibody against a protein of interest. | Enrichment of genomic sequences over which a specific protein associates in vivo. | [8] |
| ChIP-Seq: Chromatin Immunoprecipitation Sequencing | Following ChIP, samples are subjected to NGS and sequence reads are mapped onto to a reference genome. | Identification of genomic sequences over which specific protein associates in vivo. | [7] |
| 3C: Chromosome Conformation Capture | Cells are treated to cross-link DNA with nearby DNA and associated proteins. Complexes are digested with a restriction enzyme and then subjected to ligation under conditions that favor intramolecular end joining of DNA. Crosslinks are reversed and quantitative PCR is conducted with a pair of primers annealing to a different restriction enzyme fragment. | Quantifies in vivo interactions between any specific pair of genomic sequences. | [10] |
| 4C: Circular Chromosome Conformation Capture | Combines 3C and NGS. Following the 3C ligation step, another cycle of restriction enzyme digestion, dilution, and ligation is performed to generate self-circularized DNA. Inverse PCR with primers to a known sequence is conducted around these circles to amplify unknown sequences. PCR products are subject to NGS. | Quantifies in vivo interactions of a specific genomic sequence with all other genomic sequences. | [52, 53] |
| Hi-C | Combines 3C and paired-end NGS. Before ligation, single-stranded DNA is filled in and marked with biotin. After ligation, DNA is sheared, precipitated with beads linked to streptavidin, ligated to oligonucleotides, and subject to paired-end NGS. | Quantifies all in vivo pairwise interactions between genomic sequences. | [9] |
| ChIP-3C (ChIP-loop) | Combines 3C with NGS. Following 3C ligation, ChIP is performed and samples are subject to NGS. | Identifies interactions between any pair of sequences mediated by a specific protein. | [54] |
| ChIA-PET: Chromatin Interaction Analysis by Paired-End Tag Sequencing | Combines Hi-C and ChIP. Hi-C is conducted following ChIP. | Detects all genomic interactions mediated by a specific protein. | [57] |
| RNA-Seq: RNA Sequencing | NGS analysis of total cellular RNA or distinct types of cellular RNAs. | Identifies and quantifies expressed RNA sequences, including normal and mutant. | [61] |
Super-enhancers and their target gene promoters typically reside on the same structural loop [20, 22]. Notably, most inherited human disease-associated genomic sequence variants of non-coding regions locate within super-enhancers of relevant diseased cell types, including cancer cells bearing super-enhancers near their relevant transcriptionally activated oncogenes [20].
As cancer is a disease of aberrant gene expression caused by the acquisition of genetic changes. It took little time for researchers to discover somatic genomic lesions that establish super-enhancers, disrupt CTCF binding sites, or otherwise, alter genomic architecture; these were recognized as key drivers of oncogenic transcription and cancer cell growth. This review summarizes three recent studies of human T-ALL cancer cells that have formed a foundation for how alterations of non-coding regions can underlie malignant cellular transformation, and highlights how these findings raise important issues to consider in developing novel diagnostic and therapeutic strategies for conquering cancer.
Ectopic Transcription of the TAL1 Proto-Oncogene Causes T-ALL
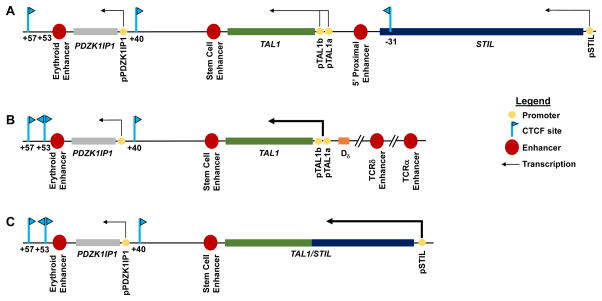
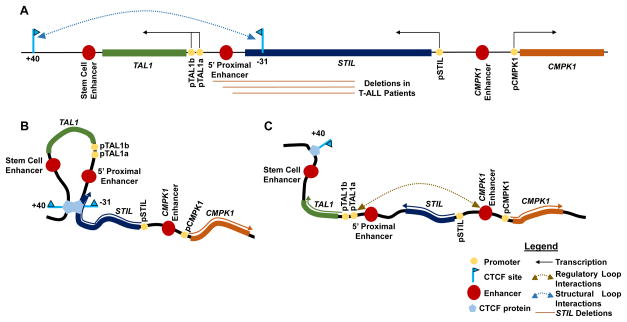
T cell acute lymphoblastic leukemia (T-ALL) is an aggressive and often fatal malignancy of immature T cells that afflicts children and adults [23]. Approximately 60% of these cancers are driven by ectopic expression of the T cell acute lymphoblastic leukemia 1 (TAL1) gene [24]. TAL1 resides at human chromosome 1p32 between PDZK1IP1 and STIL, with all three genes laying in the same transcriptional orientation (Figure 1A). TAL1 is transcribed in hematopoietic stem cells (HSC) and in erythroid and myeloid -- but not in lymphoid -- lineage hematopoietic cells, [25, 26]. PDXK1IP1 is co-expressed with TAL1, whereas STIL is ubiquitously transcribed [27]. In ~5% of patients with TAL1-expressing (TAL1+) T-ALL cancer cells, clonal chromosome t(1;14)(p32;q11) translocations fuse the T cell antigen receptor α/δ (TCRAD) locus near TAL1 drive TAL1 transcription, presumably through TCRAD locus cis-regulatory elements active in T lymphoid cells (Figure 1B) [28–31]. In ~30% of patients with TAL1+ T-ALL cells, clonal interstitial chromosome deletions with breakpoints mapping the 5′ end of TAL1, and within STIL (the TAL1d lesion), generate STIL/TAL1 fusion transcripts driven by the STIL promoter (Figure 1C) [32–34]. Studies demonstrating that ectopic Tal1 transcription in mouse T lymphoid cells causes lymphoma/leukemia supports the idea that TAL1/TCRAD genomic lesions cause T-ALL by activating TAL1 transcription in immature T lymphoid cells[35, 36]. Notably, in ~60% of patients with TAL1+ T-ALL, ectopic monoallelic TAL1 transcription occurs in the absence of detectable gross genomic alterations of the TAL1 locus [37, 38]. The pathogenesis of these cancers had remained unknown until the recent application of genome-wide technologies, as outlined below.
Figure 1.
Genomic Organization of TAL1 Loci in Normal and T-ALL Cells. (A) In normal cells, TAL1 resides between PDZK1IP1 and STIL. Indicated are the promoters and transcriptional orientations of all three genes, known TAL1 enhancers, and relevant CTCF-binding sites with their orientation indicated by arrowheads. (B) In ~5% of patients with TAL1+ T-ALLs, one allele of the TAL1 locus harbors a translocation that positions the TCRAD locus and its promoters/enhancers near the TAL1 gene. (C) In ~25% of patients with TAL1+ T-ALLs, one allele of the TAL1 locus contains an interstitial deletion that places TAL1 under control of the STIL promoter.
TAL1 Transcription in Normal Cells is Directed at Multiple Levels
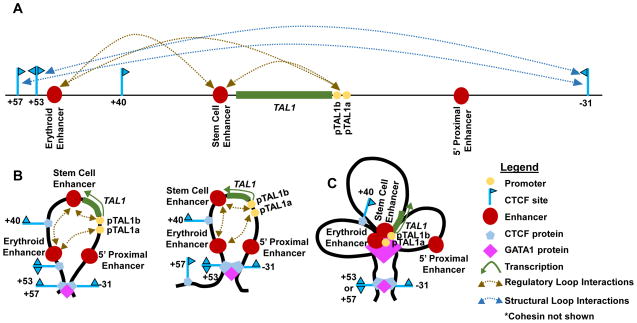
Discoveries of TAL1 as a major oncogene in T-ALL, and as an essential protein for hematopoiesis [25, 26], prompted investigations into the molecular mechanisms governing normal TAL1 transcription. Such studies examined the PDZK1IP1-TAL1-STIL loci of humans and mice since their transcription patterns, genomic composition, and known cis-regulatory elements are conserved [39]. Some studies analyzed TAL1+ K562 human erthyroleukemia and TAL1− HBP-ALL human lymphoid cell lines, with confirmation of key data in primary human and mouse cells [40, 41]. Two promoters (comprising 1a and 1b, and collectively termed the TAL1 promoter), in addition to three enhancers (erythroid, stem cell, and 5′ proximal) (Figure 2A) orchestrate transcription of TAL1 in different hematopoietic cell types [39, 42–46]. Focused 3C analyses have shown that the TAL1 promoter interacts with erythroid and stem cell enhancers in TAL1+ erythroid cells (Figure 2A) [40, 41], at higher frequencies than in TAL1− lymphoid cells [40]. The GATA-binding factor 1 (GATA1) transcription factor is an essential regulator of erythroid cell development and function, but is not expressed in lymphoid cells [44]. In erythroid cells, GATA1 activates TAL1 transcription by binding the TAL1 promoters as well as erythroid and stem cell enhancers, forming regulatory loops among these cis elements (Figure 2B) [41]. In addition, the methylation of lysine 4 on histone H3 in chromatin (forming H3K4me2 or H3K4me3) correlates with enhancer/promoter activities and regulatory loops [47, 48]. The ubiquitously-expressed hSET1 methyltransferase is the only enzyme known to methylate H3K4. In TAL1+ HSCs and erythroid precursor cells, hSET1 binding, H3K4me2, and H3K4me3 all are enriched at the TAL1 promoter and erythroid enhancer[40]. Indeed, knock-down of hSET1 protein expression in K562 cells leads to reduced TAL1 transcription, loss of regulatory loops between the TAL1 promoter and erythroid enhancer, lower histone H3 lysine 4 methylation and polymerase PolII binding at these cis-regulatory elements [39]. Collectively, these experiments demonstrate essential roles for the GATA1 and hSET1 proteins in directing normal TAL1 transcription in erythroid cells.
Figure 2.
TAL1 Locus Architecture in TAL1+ Erythroid Cells. (A) Interactions detected by 3C between cis-regulatory elements or between CTCF-binding sites are shown. (B) Each of the deduced GATA1-dependent structural loops with contacts between cis-regulatory elements are shown. (C) GATA-dependent structural loops and regulatory loops associated with active TAL1 transcription are shown
Flanking TAL1 are four CTCF-binding sites (+57, +53, +40, and −31 relative to the TAL1 transcription start site or TSS)(Figure 2A); each are constitutively bound by CTCF/cohesin and possess intrinsic enhancer-blocking activities [27, 42, 49]. Computational analysis indicates that all of these CTCF sites toward TAL1 (Figure 2A). The convergent orientation of each pair of CTCF-binding sites flanking TAL1 predicts that each pair could establish a structural loop.
Indeed, directed 3C analyses have shown that interactions between the +57 and −31 sites and between the +53 and −31 sites occur in erythroid cells (Figure 2A) [40, 41], at greater frequencies than in lymphoid cells [41]. Moroever, GATA1 protein expression promotes these two CTCF-binding site interactions, presumably to compartmentalize the TAL1 promoter, as well as the erythroid and stem cell enhancers, on a ~88 kilobase (kb) structural loop that facilitates functional interactions among these cis-regulatory elements (Figure 2B) [41]. In contrast, expression of hSET1 is not required for these two interactions, indicating that hSET1 requires pre-existing structural loops to drive TAL1 transcription [40]. These observations are consistent with the GATA1-dependent, transcription-independent assembly of a structural loop in erythroid cells that facilitates the formation of promoter/enhancer regulatory loops required to activate TAL1 expression (Figure 2C) [41]. Consequently, the absence of GATA1 protein expression in T lymphoid cells may be critical to prevent TAL1 transcription, thereby suppressing the occurrence of T-ALL. Although several proteins, cis elements, and 3D chromosome structures that direct TAL1 transcription in erythroid cells have been identified, the complete array of molecular mechanisms by which these and additional factors promote TAL1 expression in hematopoietic cells remains to be elucidated. Extrapolating existing and newer findings from erythroid cells in the future might help provide information on the precise mechanistic events occurring within immature T cells that lead to T-ALL.
Activation of TAL1 Transcription by Genomic Alterations of Non-Coding Regions
In the past two years, three independent studies using genome-wide analyses have reported previously unappreciated mechanisms that stimulate transcription of TAL1 in T-ALL cells lacking known genomic alterations within the TAL1 locus. A common model for these studies is the TAL1+ Jurkat human T-ALL cell line that displays monoallelic TAL1 transcription in the absence of TAL1d or TAL1/TCRAD lesions [40, 50, 51].
An acquired interchromosomal interaction between cis-regulatory elements activates TAL1
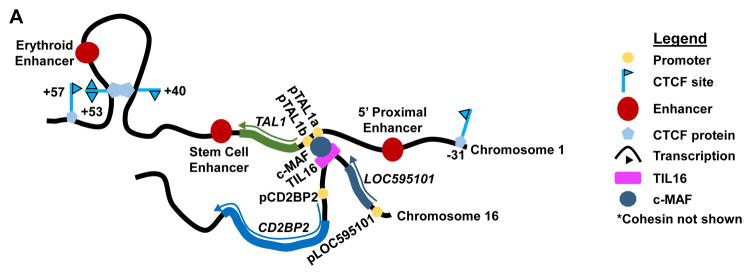
In 2014, the Huang lab reported that an interchromosomal interaction between a T cell enhancer and the TAL1 promoter could activate transcription of TAL1 in Jurkat and other TAL1+ T-ALL cells lacking TAL1d or TAL1/TCRAD lesions [40]. ChIP-Seq revealed the enrichment of hSET1 and H3K4 methylation at TAL1 promoters, but not enhancers, in the Jurkat and Rex T-ALL cell lines. Consistent with this finding, the erythroid enhancer neither contacted the TAL1 promoter nor stimulated a luciferase reporter gene in Jurkat or Rex cells. Directed 3C analyses of the TAL1 locus revealed that Jurkat and TAL1+ primary T-ALL cells lacked the +53/−31 CTCF-binding site interaction of erythroid cells, yet contained a +53/+40 CTCF site interaction (Figure 3A) not present in erythroid cells (Figure 2A). These data suggest that the cis-regulatory elements and 3D chromosome structures that direct TAL1 transcription in erythroid cells do not activate TAL1 transcription in T-ALL cells. The +53/+40 CTCF site interaction is also not present in the TAL1− HBP-ALL T-ALL cell line implying that TAL1 adopts a unique 3D chromosome structure in TAL1+ T-ALL cells. Analysis of Jurkat cells by 4C (Table 1) [52, 53], which couples NGS with 3C to determine all genomic interactions for a particular sequence (Table 1), revealed that the TAL1 promoter contacts a non-coding region of chromosome 16 (Figure 3B), named TIL16 for TAL1-interacting locus located in chromosome 16 [40]. TIL16 lies between the long non-coding RNA LOC595101 locus and the T cell signaling factor CD2BP2 locus (Figure 3B), which are both transcribed in T cells. Subsequent 3C assays confirmed this interaction in Jurkat cells, as well as other TAL1+ T-ALL cell lines and primary T-ALL cells lacking TAL1d or TAL1/TCRAD. TIL16 contains binding sites for many transcription factors including the T cell specific c-MAF proto-oncogene. The ability of TIL16, but not a c-MAF-binding site-inactivated TIL16, to activate a TAL1 promoter-driven reporter in Jurkat cells showed that TIL16 was actually an enhancer. In Jurkat cells, c-MAF knock-down caused loss of the TAL1/TIL16 contact, decreased TAL1 mRNA expression, and reduced cellular proliferation. Moreover, ChIP-3C (or ChIP-loop) [54], which couples NGS with 3C to identify genomic interactions bridged by a protein (Table 1), showed that c-MAF formed a complex with TIL16 and the TAL1 promoter (Figure 3B), which also contained a c-MAF binding site. These data are consistent with a model wherein aberrant formation of a c-MAF-dependent interchromosomal regulatory loop allows the TIL16 enhancer to drive oncogenic TAL1 transcription. Consequently, this provides a substantial conceptual advance regarding how aberrant interactions between cis-regulatory elements on different chromosomes can activate oncogenic transcription of a proto-oncogene. Namely, epigenetic or genetic changes enable a transcription factor to bridge functional communications between an enhancer on one chromosome and a proto-oncogene promoter on another chromosome, thereby driving malignant cellular transformation. However, the mechanistic basis for how such interchromosomal contacts are acquired, remains to be determined.
Figure 3.
An Interchromosomal Interaction Drives TAL1 Transcription in T-ALL Cells. Representation of the contact detected by 4C between the TAL1 promoter on chromosome 1 and the TIL16 enhancer on chromosome 16. This interchromosomal interaction depends on expression of the c-MAF transcription factor, which binds to sequences within both TIL16 and the TAL1 promoter. An intrachromsomal contact between CTCF sites downstream of TAL1 is observed by 3C. This presumably represents a CTCF-mediated structural loop.
Creation of a TAL1 locus super-enhancer by somatic mutation activates TAL1 transcription
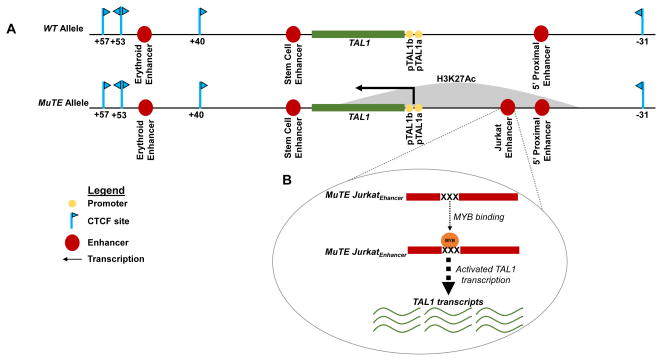
Later in 2014, the Look lab discovered that somatic acquisition of insertions in a non-coding region near TAL1 established a super-enhancer that activated TAL1 transcription in T-ALL cells [51]. The goal of their study was to assess if genomic alterations affecting cis-regulatory elements could activate monoallelic TAL1 transcription in TAL1+ T-ALL cells lacking TAL1d or TAL1/TCRAD lesions. ChIP-Seq revealed aberrantly high density and breadth of histone H3 lysine 27 acetylation (H3K27Ac) from −20 kb through +10 kb of the TAL1 TSS in Jurkat cells (Figure 4), but not in other T-ALL cell lines or normal human hematopoietic stem cells. Moroever, in 2013, the Vetrie lab had analyzed Jurkat cells by 3C, finding contacts between the TAL1 promoter and sequences −8 or −10 kb of the TAL1 TSS (Figure 4) [41]. These sequences were actually known to correspond to the TAL1 5′ proximal enhancer (−10 kb) and “Jurkat enhancer” (−8 kb), a cis-regulatory element that TAL1 and other transcription factors bind, to promote TAL1 transcription in Jurkat cells (Figure 4) [55]. Genomic sequencing of Jurkat cells indicated a monoallelic 12 (base pair) bp insertion at sequences within the −8 kb TAL1 ChIP-Seq peak. Monoallelic insertions of 2–18 bp were found in one out of eight other TALL lines (Molt3) and eight out of 146 primary T-ALL samples, but not in normal cells (controls) from two of the T-ALL patients. These sequence data indicate that “mutation of the TAL1 enhancer” (MuTE) can be acquired in immature T cells, or in cells that differentiate into the T lymphoid lineage. The H3K27Ac profile of the Jurkat MuTE suggested that this genomic alteration might create a TAL1 super-enhancer [51]. Consistent with this idea, TAL1 mRNA was expressed from only MuTE alleles in T-ALL cells from five patients with allele-specific polymorphisms in their TAL1 3′ UTRs [51]. Moreover, each MuTE insertion created at least one predicted binding site for the MYB transcription factor (Figure 4). Genomic fragments spanning MuTE insertions activated the expression of a luciferase reporter gene in Jurkat cells, while MYB knock-down lowered the reporter activity. ChIP-Seq demonstrated that MYB could bind to the MuTE insertions in Jurkat and Molt3 cells (Figure 4), but not to corresponding “normal” regions in TAL− T-ALL cell lines, or a primary TAL1d T-ALL. ChIP-Seq reads from Jurkat and Molt3 cells further indicated that MYB bound −8 kb of the TAL1 TSS on only MuTE alleles (Figure 4), highlighting that MuTE establishes a cis-regulatory element that drives oncogenic TAL1 transcription. Of note, an attempt to delete the MuTE insertion in Jurkat cells using CRISPR/Cas9 genomic editing [56], proved unsuccessful due to problems expanding targeted clones, and presumably reflecting the dependence of Jurkat cellular proliferation on TAL1 protein expression. Consistent with this notion, retrovirus-driven TAL1 expression has enabled the isolation of MuTE-deleted Jurkat lines, which express lower levels of endogenous TAL1 mRNA relative to parental Jurkat cells, demonstrating that MuTE is the causative lesion that drives oncogenic TAL1 transcription. Indeed, ChIP-Seq of these lines has shown loss of MYB binding and H3K27Ac at the TAL1 locus, demonstrating that the Jurkat TAL1 super-enhancer requires the MuTE-inserted MYB binding sites for efficient TAL1 transcription. This study thus established the paradigm that somatic mutation of a non-coding region can introduce binding sites for a transcription factor. The binding of the transcription factor in turn, establishes a super-enhancer, and this is significant because it can then activate oncogenic transcription of a nearby proto-oncogene.
Figure 4.
An Acquired Super-Enhancer Activates Monoallelic TAL1 Transcription in T-ALL Cells. (A) Depictions of the TAL1 loci on the wild-type (WT) and MuTE alleles in T-ALL cells showing the location of the MuTE super-enhancer detected by ChIP-Seq for H3K27Ac. (B) The MuTE insertions create bindings sites for MYB, establishing a super-enhancer which then activates monoallelic transcription of TAL1, as determined by genomic sequencing, ChIP-Seq, and RNA-Seq.
Deletion of a CTCF-binding site alters TAL1 architecture and activates TAL1 transcription
In 2016, the Young lab discovered that deletions spanning CTCF-binding sites which compartmentalized proto-oncogenes within chromosome structural loops, activated oncogenic transcription of these genes in human T-ALL cells [50]. The group had previously shown that intra-TAD structural loops created insulated neighborhoods important for proper transcriptional regulation of their composite genes [22]. They sought to test the hypothesis that disruption of insulated neighborhoods containing repressed proto-oncogenes might enable active enhancers from adjacent neighborhoods to induce oncogenic transcription of these genes. The analysis of Jurkat cells by ChIA-PET [57], a technique combining ChIP-Seq and 3C approaches (Table 1), identified ~9,000 CTCF/cohesin-mediated chromosome structural loops (insulated neighborhoods). The transcribed TAL1 gene and the Jurkat TAL1 super-enhancer were found within an insulated neighborhood whose borders constituted the TAL1 locus +40 and −31 CTCF-binding sites (Figure 5A and 5B). Analysis of sequence data from primary T-ALL cells identified genomic deletions spanning the −31 site (residing within STIL), but not extending into the TAL1 promoter, in contrast to TAL1d lesions, which do (Figure 5A). This suggested that disruption of the TAL1 insulated neighborhood might cause oncogenic TAL1 transcription. One prediction of this model was that deletion of the −31 CTCF site in cells where TAL1 is repressed would activate TAL1 transcription by enabling an enhancer in the adjacent structural loop to interact with the TAL1 promoter. Indeed, CRISPR/Cas9-mediated deletion of 400 bp spanning this CTCF site in human embryonic kidney cells or primary human T cells resulted in higher levels of TAL1 transcripts (Figure 5C). This deletion was also found to permit interactions between sequences normally compartmentalized within the TAL1 or adjacent insulated neighborhood (Figure 5C) [50]. In parallel to these TAL1 analyses, the study showed that Jurkat and primary T-ALL cells harbored deletions of CTCF-binding sites disrupting the border of the LIM Domain Only 2 (LMO2) proto-oncogene insulated neighborhood. This in turn, stimulated LMO2 transcription. Moreover, data from the International Cancer Genome Consortium indicated that a greater frequency of somatic mutations existed in CTCF-binding sites marking insulated neighborhood boundaries relative to other non-coding regions of human esophageal and liver cancer cell genomes [50]. Notably, in some instances, these mutations lie within the border of an insulated chromosome neighborhood containing a proto-oncogene whose transcriptional activation causes the relevant malignancy. Thus, this study established the paradigm that somatic deletions within non-coding regions can disrupt borders of insulated neighborhoods containing silent proto-oncogenes, thereby activating their transcription, which under normal circumstances, would be repressed.
Figure 5.
Disruption of the TAL1 Insulated Neighborhood Border Activates TAL1 Transcription in T-ALL Cells. (A) Representation of the predominant cohesin-mediated structural loop identified by ChIA-PET analysis of Jurkat T cells. (B) Diagram of the TAL1 insulated neighborhood in normal cells. This structural loop isolates the TAL1 promoters from enhancers outside of this structural loop. (C) Diagram of the TAL1 locus configuration, interactions, and transcriptional status in HEK-293T cells following deletion of the TAL1 −31 CTCF-binding site. The deletion of this sequence disrupts the normal structural loop and enables distal enhancers to aberrantly communicate with the TAL1 promoter to drive oncogenic TAL1 expression.
Fundamental issues raised by genome-wide studies of T-ALL cells
These studies raise fundamental issues regarding the discovered genomic aberrations of non-coding regions and human T-ALL pathogenesis. A central issue is the extent to which each aberration drives malignant transformation of immature T cells. A related issue is whether any of these aberrations can cooperate to cause T-ALL via enhanced activation of TAL1 transcription. While the establishment and analysis of mice with T cell-specific conditional activation of each genetic abnormality alone, or in combination with others, might provide answers for developing T cells, this approach is feasible only for deletion of the TAL1 −31 CTCF-binding site using current technology. However, for this particular genomic alteration in addition to MuTE, mice carrying analogous lesions alone, or together in the germline, could provide first approximations.
Of note, a recent study of human gliomas showed that mutation of a metabolic enzyme could indirectly activate a key proto-oncogene through increased DNA methylation, which in turn antagonized CTCF binding to disrupt TAD borders [58]. It will be important to determine whether similar pathogenic mechanisms cause oncogenic transcription of proto-oncogenes in T-ALL. Another issue is how T-ALL cells acquire interchromosomal contacts between the TAL1 promoter and TIL16 enhancer. Possibilities include aberrant activation of the c-MAF proto-oncogene, establishment of a TIL16 super-enhancer, and disruption of intra-chromosome structural loops to limit contacts between cis-regulatory elements on chromosomes 1 and 16. Regardless of the underlying mechanisms, it would be valuable to assess the extent to which the c-MAF oncogene causes T lineage lymphoma/leukemia by driving ectopic TAL1 transcription. Finally, considering that an alternative TAL1 promoter (IV) can be activated in T-ALL cells lacking the TAL1d or TAL1/TCRAD oncogenic lesions [59], the roles of TIL16, MuTE, TAL1 CTCF-binding site deletions, and possibly other non-coding region alterations in activating the TAL1 promoter IV warrant investigation.
Another important issue to consider is why the 4C analysis performed on Jurkat cells by the Huang lab identified the TAL1 promoter interacting with TIL16, but not the 5′ proximal and “Jurkat cell” enhancers [40]. The Vetrie lab previously identified interactions of the TAL1 promoter with these latter two enhancers by conducting biased 3C analyses on Jurkat cells [41]. One possible explanation is that the small number of 4C sequence reads analyzed was insufficient to detect TAL1 promoter interactions with the TAL1 locus enhancers and that these interactions, for example, might occur less often than TAL1 promoter contacts with the TIL16 enhancer. Another possibility is that c-MAF-mediated bridging of the TAL1 promoter and TIL16 is very stable and therefore, less prone to dissolution during sample preparation. Alternatively, genetic and/or epigenetic differences between the Jurkat cells analyzed might confer distinct 3D chromosome structures and interactions upon the TA1L locus. Since being published in 1977 [60], the Jurkat cell line has been widely and repeatedly distributed by multiple sources. Moreover, different derivatives of the Jurkat cell line have been established. As TAL1 expression drives proliferation of Jurkat cells, it is conceivable that the splitting and passaging of these cells has resulted in sub-lines with acquisition and/or dependence on different mechanisms for sustaining high-level TAL1 transcription. Regardless, the identification of contacts between the TAL1 promoter and the TIL16 enhancer in primary human T-ALL samples confirm that this aberrant interchromosomal interaction occurs in vivo.
Concluding Remarks
Recent discoveries that genomic alterations of non-coding regions activate ectopic transcription of the TAL1 proto-oncogene in T-ALL have implications for the clinical management of patients with this aggressive and often fatal cancer (Box 1). Moreover, since these pathogenic mechanisms are certainly relevant for other and likely all human cancers, the lessons from T-ALL should be universally applicable. A mainstay of cancer therapeutics is the development and administration of drugs that selectively inhibit key oncogenes driving malignant cellular growth in each patient’s cancer cells. Shortcomings of this strategy include difficulties in identifying key oncogenes and developing drugs that selectively target these, as well as identifying the acquisition of mutations that render these oncogenes insensitive to drug inhibition. Overcoming these challenges requires continuous advances in our basic understanding of how proto-oncogenes function in normal cells, on the genomic alterations that induce oncogenic activities of proto-oncogenes, and on the resultant oncogenes that drive malignant cellular transformation (see Outstanding Questions). Ideally, in order to design personalized therapies, diagnostic characterizations of cancer cells would include genome-wide analyses to identify all mutated proto-oncogenes, over-expressed proto-oncogenes, novel super-enhancers, aberrant interactions among cis-regulatory elements, and disrupted boundaries of insulated neighborhoods or TADs. However, such comprehensive analyses are not practical today due to costs and limitations of technologies, in addition to our superficial knowledge of how genomic non-coding regions regulate gene transcription. Until advances in basic science remove these obstacles, one conservative path forward would be to perform whole exome analyses via RNA-Seq (Table 1), in order to identify all mutated and/or over-expressed proto-oncogenes [61]. For over-expressed proto-oncogenes, subsequent assays for a linked super-enhancer (via H3K27Ac ChIP-Seq) [62] or for determining new contacts of respective promoters with their enhancers (via 4C), could be employed to identify factors driving increased transcription. This knowledge might enable oncologists to develop or use drugs targeting relevant oncogenes. Furthermore, these might prove to be promising in combination with other drugs that selectively inhibit super-enhancers [63, 64] or classical enhancers, which are critical for the expression of key oncogenes.
Box 1. The Clinician’s Corner.
Ectopic expression of the T cell acute lymphoblastic leukemia 1 (TAL1) proto-oncogene occurs in the majority of T-ALL cancers.
TAL1 is transcriptionally silent and resides on a structural loop in normal immature T cells.
The acquisition of an abnormal interaction between the TAL1 promoter and an enhancer on a distinct chromosome can activate the TAL1 promoter and promote ectopic TAL1 expression in T-ALL cells.
The acquisition of a mutation within the TAL1 structural loop can create a super-enhancer that activates the TAL1 promoter and drives ectopic TAL1 expression in T-ALL cells.
The deletion of genomic sequences spanning one of the CTCF-binding sites that defines the TAL1 structural loop enables an enhancer on an adjacent structural loop to activate the TAL1 promoter and drive ectopic TAL1 expression in T-ALL cells.
Outstanding Questions.
What genetic and/or epigenetic changes promote abnormal interchromosomal communication between an active enhancer on one chromosome and a proto-oncogene promoter on the other chromosome?
Do genetic lesions that create super-enhancers or disrupt insulated neighborhood boundaries arise spontaneously? Alternatively, do they arise from mutations in genes encoding DNA replication or repair factors?
Are super-enhancers quantitatively and/or qualitatively different than classical enhancers?
What are the mechanisms by which super-enhancers form and function?
What is the full menu of noncoding genomic region alterations that activate gene transcription changes to drive the development, progression, drug-resistance, and relapse of T-ALL?
What are the mechanistic bases by which each of these genomic alterations influences T-ALL pathogenesis?
Trends.
Recent advances in genome-wide technologies are promoting unprecedented increases in our understanding of how non-coding regions of the human genome regulate gene transcription.
The TAL1 proto-oncogene, associated with T-ALL malignancies, resides within a genomic structural “insulated neighborhood” that is active in many hematopoietic lineage cells but normally repressed in lymphocytes.
Abnormal interchromosomal contacts between an enhancer on chromosome 16 and the TAL1 promoter on chromosome 1 stimulate TAL1 transcription in T-ALL cells.
Somatic mutations that establish a novel super-enhancer overlapping the TAL1 promoter drive TAL1 transcription in T-ALL cells.
Deletions disrupting a border of the TAL1 insulated neighborhood activate TAL1 transcription in immature T cells, thereby causing T-ALL.
Acknowledgments
The work on this review was supported by the 5T32GM007229 Training Program in Cell and Molecular Biology of the University of Pennsylvania (A.R.-R.), as well as Pennsylvania Tobacco Settlement Funds and NIH R01 grant AI112621 (C.H.B).
Glossary
- CCCTC-binding factor (CTCF) chromosome structural protein
a ubiquitously expressed DNA binding protein that forms structural chromosome loops, and which organizes three-dimensional genomic architecture.
- ChIP-Seq peak
a region of the genome to which numerous sequence reads of a ChIP-Seq experiment map.
- Chromatin immuneprecipitation-sequencing (ChIP-Seq)
a method employed to identify all genomic sequences to which a specific protein associates.
- Chromosome loops
topological units of genomes established by interactions between proteins bound at specific sequences.
- Cis-acting factors
DNA sequences that regulate transcription of genes on the same chromosome.
- Clonal interstitial chromosome deletions
the loss of internal chromosome sequences in every cell within a population.
- Cohesin proteins
multi-protein subunit complexes that function with CTCF to establish chromosome loops.
- CRISPR/Cas9 genomic editing
a methodology that uses small RNAs to guide bacterial nucleases to specific genomic sites, and uses homologous sequences with a defined mutation to repair broken DNA and introduce the mutation.
- CTCF binding site
specific DNA sequence to which CTCF binds.
- Enhancers
DNA sequences that activate promoters.
- Insulated neighborhoods
chromosome structural loops whose gene promoters are protected from outside enhancers.
- Next generation sequencing (NGS)
high-throughput sequencing methodologies that involve massive parallelization of the sequencing process to produce thousands or millions of sequences concurrently.
- Proto-oncogene
a gene that can become an oncogene through mutation and/or overexpression.
- Regulatory loops
chromosome loops formed by interactions between proteins bound to promoters and enhancers.
- Somatic mutation
a genetic alteration acquired in a somatic cell.
- Structural loops
chromosome loops formed by interactions between CTCF/cohesin complexes bound at CTCF-binding sites.
- Super-enhancers
non-coding genomic regions that contain multiple classical enhancers and exhibit robust ability to activate transcription from promoters.
- T-cell acute lymphoblastic leukemia (T-ALL)
an aggressive clonal malignancy of immature T cells that arises in children and adults.
- Topological association domains (TADs)
megabase-sized genomic regions within which sequences interact more frequently than with sequences outside of these regions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 2.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sur I, et al. Lessons from functional analysis of genome-wide association studies. Cancer Res. 2013;73(14):4180–4. doi: 10.1158/0008-5472.CAN-13-0789. [DOI] [PubMed] [Google Scholar]
- 4.Oldridge DA, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528(7582):418–21. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melton C, et al. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet. 2015;47(7):710–6. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katainen R, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47(7):818–21. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 8.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–31. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dekker J, et al. Capturing chromosome conformation. Science. 2002;295(5558):1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 11.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–6. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14(6):762–75. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160(6):1049–59. doi: 10.1016/j.cell.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 15.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30(8):881–91. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupianez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–25. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groschel S, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369–81. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511(7510):428–34. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110(44):17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159(2):374–87. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3398–406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 25.Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86(1):47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 26.Robb L, et al. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 1996;15(16):4123–9. [PMC free article] [PubMed] [Google Scholar]
- 27.Delabesse E, et al. Transcriptional regulation of the SCL locus: identification of an enhancer that targets the primitive erythroid lineage in vivo. Mol Cell Biol. 2005;25(12):5215–25. doi: 10.1128/MCB.25.12.5215-5225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Begley CG, et al. Chromosomal translocation in a human leukemic stem-cell line disrupts the T-cell antigen receptor delta-chain diversity region and results in a previously unreported fusion transcript. Proc Natl Acad Sci U S A. 1989;86(6):2031–5. doi: 10.1073/pnas.86.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard O, et al. Two distinct mechanisms for the SCL gene activation in the t(1;14) translocation of T-cell leukemias. Genes Chromosomes Cancer. 1990;1(3):194–208. doi: 10.1002/gcc.2870010303. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, et al. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. EMBO J. 1990;9(2):415–24. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finger LR, et al. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc Natl Acad Sci U S A. 1989;86(13):5039–43. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll AJ, et al. The t(1;14)(p34;q11) is nonrandom and restricted to T-cell acute lymphoblastic leukemia: a Pediatric Oncology Group study. Blood. 1990;76(6):1220–4. [PubMed] [Google Scholar]
- 33.Breit TM, et al. Site-specific deletions involving the tal-1 and sil genes are restricted to cells of the T cell receptor alpha/beta lineage: T cell receptor delta gene deletion mechanism affects multiple genes. J Exp Med. 1993;177(4):965–77. doi: 10.1084/jem.177.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown L, et al. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J. 1990;9(10):3343–51. doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Condorelli GL, et al. T-cell-directed TAL-1 expression induces T-cell malignancies in transgenic mice. Cancer Res. 1996;56(22):5113–9. [PubMed] [Google Scholar]
- 36.Kelliher MA, Seldin DC, Leder P. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIalpha. EMBO J. 1996;15(19):5160–6. [PMC free article] [PubMed] [Google Scholar]
- 37.Leroy-Viard K, et al. Distinct DNase-I hypersensitive sites are associated with TAL-1 transcription in erythroid and T-cell lines. Blood. 1994;84(11):3819–27. [PubMed] [Google Scholar]
- 38.Ferrando AA, et al. Biallelic transcriptional activation of oncogenic transcription factors in T-cell acute lymphoblastic leukemia. Blood. 2004;103(5):1909–11. doi: 10.1182/blood-2003-07-2577. [DOI] [PubMed] [Google Scholar]
- 39.Gottgens B, et al. Long-range comparison of human and mouse SCL loci: localized regions of sensitivity to restriction endonucleases correspond precisely with peaks of conserved noncoding sequences. Genome Res. 2001;11(1):87–97. doi: 10.1101/gr.153001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel B, et al. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia. 2014;28(2):349–61. doi: 10.1038/leu.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. Chromatin looping defines expression of TAL1, its flanking genes, and regulation in T-ALL. Blood. 2013;122(26):4199–209. doi: 10.1182/blood-2013-02-483875. [DOI] [PubMed] [Google Scholar]
- 42.Dhami P, et al. Genomic approaches uncover increasing complexities in the regulatory landscape at the human SCL (TAL1) locus. PLoS One. 2010;5(2):e9059. doi: 10.1371/journal.pone.0009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aplan PD, et al. The SCL gene is formed from a transcriptionally complex locus. Mol Cell Biol. 1990;10(12):6426–35. doi: 10.1128/mcb.10.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogilvy S, et al. The SCL +40 enhancer targets the midbrain together with primitive and definitive hematopoiesis and is regulated by SCL and GATA proteins. Mol Cell Biol. 2007;27(20):7206–19. doi: 10.1128/MCB.00931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottgens B, et al. The scl +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5′ bifunctional hematopoietic-endothelial enhancer bound by Fli-1 and Elf-1. Mol Cell Biol. 2004;24(5):1870–83. doi: 10.1128/MCB.24.5.1870-1883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bockamp EO, et al. Lineage-restricted regulation of the murine SCL/TAL-1 promoter. Blood. 1995;86(4):1502–14. [PubMed] [Google Scholar]
- 47.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 49.Follows GA, et al. Mapping and functional characterisation of a CTCF-dependent insulator element at the 3′ border of the murine Scl transcriptional domain. PLoS One. 2012;7(3):e31484. doi: 10.1371/journal.pone.0031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hnisz D, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351(6280):1454–8. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346(6215):1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38(11):1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38(11):1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 54.Horike S, et al. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37(1):31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 55.Sanda T, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):209–21. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–4. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernard O, et al. A third tal-1 promoter is specifically used in human T cell leukemias. J Exp Med. 1992;176(4):919–25. doi: 10.1084/jem.176.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider U, Schwenk HU, Bornkamm G. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer. 1977;19(5):621–6. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- 61.Morin R, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45(1):81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 62.Vaharautio A, Taipale J. Cancer. Cancer by super-enhancer. Science. 2014;346(6215):1291–2. doi: 10.1126/science.aaa3247. [DOI] [PubMed] [Google Scholar]
- 63.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159(5):1126–39. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]