Abstract
Cell based assessment tools and screening platforms are the preferred paradigm for small molecule identification and validation due to selectively identifying molecules with cellular activity and validation of compound activity against target proteins in their native environment. With respect to Regulator of G Protein Signaling (RGS) proteins, current cell based methodologies are either low throughput or monitor downstream signaling consequences. The increasing number of reports indicating RGS function in various disease pathogeneses highlights the need for a robust RGS inhibitor discovery and characterization paradigm. Promega’s NanoBit Protein Complementation Assay utilizes NanoLuc, an engineered luciferase with enhanced luminescence characteristics which allow for both robust and kinetic assessment of protein interaction formation and disruption. Here we characterized 15 separate RGS: G protein interactions using this system. The binding profile of RGS: Gα interactions correlates to prior published biochemical binding profiles of these proteins. Additionally, we demonstrated this system is suitable for high throughput screening efforts via calculation of Z-factors for three of the interactions and demonstration that a known small molecule inhibitor of RGS4 disrupts the RGS4: Gαi1 protein-protein interaction. In conclusion, the NanoBit Protein Complementation Assay holds promise as a robust platform for discovery and characterization of RGS inhibitors.
Keywords: NanoBit, RGS, high throughput screening, Gαi1, Gαq
Introduction
G protein-coupled receptors (GPCRs) are responsible for the initiation of a wide range of cellular signaling processes in response to stimuli. These processes propagate signaling events through both heterotrimeric guanine nucleotide binding proteins (G proteins) and β-arrestins (Wisler, Xiao et al. 2014). Regulator of G protein Signaling (RGS) proteins temporally regulate heterotrimeric G protein signaling cascades elicited by GPCR activation by acting as GTPase accelerating proteins (GAPs) for active, GTP-bound Gαi/o and Gαq proteins. Formation of a protein protein interaction between RGS and Gα stabilizes the Gα transition state, accelerating cleavage of the γ phosphate of GTP and returning Gα to the GDP-bound, inactive state (Tesmer, Berman et al. 1997). As modulators of G protein signaling, RGS proteins have been implicated in a number of disease states that involve GPCR signaling. RGS6, for example, has been implicated in anxiety and depression (Stewart, Maity et al. 2014), RGS4 plays a role in Parkinson’s Disease and pain (Garnier, Zaratin et al. 2003, Lerner and Kreitzer 2012), and RGS17 overexpression can support survival of lung, prostate, breast, and hepatocellular carcinomas (James, Lu et al. 2009, Sokolov, Iannitti et al. 2011, Li, Li et al. 2015). Some RGS proteins have been implicated in pathologies due to interactions with proteins other than Gα. RGS6 plays a role in doxorubicin mediated cardiotoxicity and cardiomyopathy, as well as cytotoxicity leading to cardiomyopathy and hepatic cirrhosis associated with chronic alcohol consumption, independent of Gα binding (Yang, Maity et al. 2013, Stewart, Maity et al. 2015). The role RGS proteins play in a variety of disease states has been recently reviewed by several groups (Hurst and Hooks 2009, Sjogren 2011, Stewart, Maity et al. 2015, Hayes and Roman 2016).
As the importance of RGS proteins in disease pathogenesis emerges, targeting of RGS proteins with small molecules has gained attention. Early efforts to identify RGS inhibitors relied on flow cytometry-based biochemical screens. These efforts ultimately led to the discovery of CCG-50014, an RGS4 selective inhibitor with nanomolar potency that acts by covalently modifying cysteine residues (Roman, Ota et al. 2009, Blazer, Zhang et al. 2011). Recent reports have shown CCG-50014’s ability to reduce neuropathic hyperalgesia in vivo (Bosier, Doyen et al. 2015) and enhance opioid-mediated analgesia in vivo (Yoon, Woo et al. 2015). Furthermore, treatment of mice with CCG-50014 analog CCG-203769 caused a reversion of raclopride induced akinesia and bradykinesia, demonstrating reduction of Parkinsonian behaviors (Blazer, Storaska et al. 2015). A growing body of evidence provides validation for targeting RGS proteins for therapeutic intervention.
Techniques used to identify small molecule inhibitors of RGS proteins have focused primarily on biochemical methods, with published assays to assess protein-protein interactions between the RGS and Gα (Proximity assays i.e. ALPHAScreen, FRET, TR-FRET, etc) as well as functional assays measuring GAP activity of the RGS (Roman, Talbot et al. 2007, Roman, Ota et al. 2009, Roof, Roman et al. 2009, Mackie and Roman 2011, Monroy, Mackie et al. 2013). The techniques developed for cell based assessment of inhibitor function or primary discovery are far fewer. Blazer et al. demonstrated the ability of CCG-50014 to inhibit RGS4’s interaction with its cognate G protein alpha subunit in cells by examining changes in sub-cellular localization of a GFP-RGS4 fusion protein upon compound treatment (Blazer, Zhang et al. 2011). A calcium signaling assay has also successfully been used in a high throughput screen for inhibitors of RGS4 (Storaska, Mei et al. 2013). Cell based screening methods have several advantages over biochemical methods, including presenting targets in their native environment, and providing a cellular activity filter on primary hit compounds – a process that avoids the discovery of biochemical inhibitors with physicochemical properties that make them membrane impermeable or otherwise inactive in a cellular context. However, use of the cell based methods described above for discovery efforts presents its own challenge, such as expensive specialized equipment and necessity to determine compound target specificity.
Development of an assay broadly applicable to all RGS proteins may be advantageous in that it could allow for the high throughput comparison of small molecule selectivity among RGS proteins in a cellular context. Unfortunately, a recurrent theme in previously identified RGS inhibitors is reactivity with cysteine residues (Roman, Ota et al. 2009, Blazer, Zhang et al. 2011, Storaska, Mei et al. 2013, Vashisth, Storaska et al. 2013), so a system that allows for the use of cysteine-less RGS mutants (or other point mutants) to investigate compounds’ mechanisms of action without the need to biochemically purify those mutant proteins would be beneficial. Finally, the method should be amenable to the interrogation of RGS: protein interactions beyond RGS: Gα interactions. For example, RGS6 belongs to the more complex R7 family of RGS proteins, a hallmark of which is the formation of an obligatory heterodimer between the G protein gamma like (GGL) domain of the R7 RGS and the atypical Gβ protein Gβ5 (Snow, Krumins et al. 1998, Snow, Betts et al. 1999). As formation of a RGS6L: Gβ5 heterodimer is essential for the stability and expression of RGS6, it has been hypothesized that a therapeutic agent targeting this interaction would act to inhibit all RGS6 mediated signaling cascades be they Gα protein dependent or independent (Chen, Eversole-Cire et al. 2003). An assay that is amenable to the assessment of multiple types of RGS: protein interactions will allow for mechanistic characterization of compound effect on multiple signaling pathways.
The NanoBit protein complementation assay (Promega, Madison, WI) (Dixon, Schwinn et al. 2016) utilizes an engineered luciferase, called NanoLuc, which boasts increased and sustained luminescence compared to other luciferase systems (Hall, Unch et al. 2012). The NanoLuc enzyme is separated into two fragments, an 18kDa large bit (LB) and a 1.3kDa small bit (SB) (Dixon, Schwinn et al. 2016). To assess a protein interaction pair, plasmids are constructed to express eight separate fusion proteins, whereby the LB and SB are located on either the N or C terminus for each protein (Figure 1). An optimal arrangement can then be determined that will allow the LB and SB to form a complete, competent, enzyme when the protein pair of interest forms a protein-protein interaction. The intrinsic affinity of the LB and SB is 190 µM, which is greater than the typical range of protein interaction affinities, so the affinity of LB for SB will not drive an interaction together i.e. if a protein interaction is detected it is due to the interaction of the proteins of interest and not an artifact of the affinity of LB for SB (Dixon, Schwinn et al. 2016). Additionally, the glow kinetics unique to the enzyme allows for a kinetic assessment of the formation and disruption of protein-protein interactions in real time (Dixon, Schwinn et al. 2016).
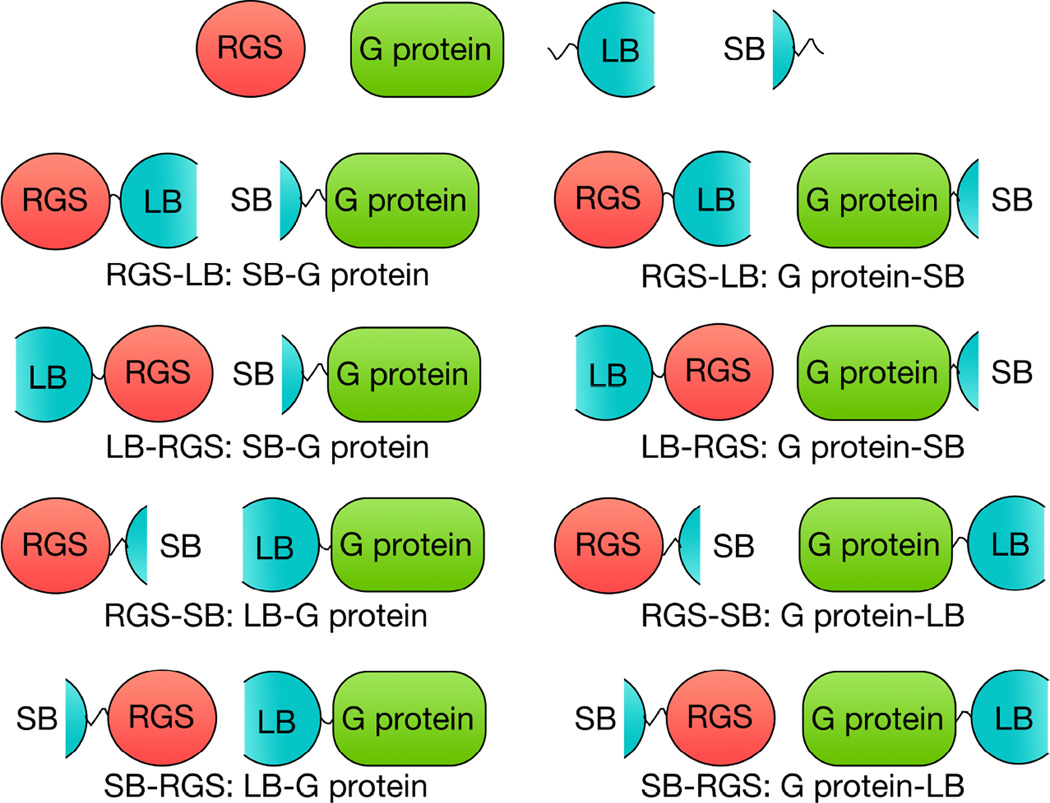
Figure 1.
Assembly of Protein Interaction Pairs. Four plasmids were constructed to produce fusion proteins of each possible large bit (LB) and small bit (SB) combination. Proteins depicted with a Nanobit piece on the left of the protein is fusion on the N-terminus of the RGS or G protein, and a Nanobit piece on the right of the protein represents fusion on the C-terminus of the RGS or G protein. Under each depiction is an example of how these fusion constructs are referred to in the text.
Here we present the development of this cellular protein complementation assay using RGS4 and RGS17 as representative RGS proteins. We also advance the development of the assay to include 10 additional RGS proteins, and one mutant RGS protein, and characterize them in this system for interactions with Gαi1 and, for RGS2, Gαq. Additionally, we demonstrate that this technology can be expanded to monitor interactions with RGS proteins and proteins other than Gα proteins. Finally, we show that this technology can be adapted for high throughput screening.
Materials and Methods
Construction of NanoBit Vectors
Human RGS1, 2, 6, 7, 10, 14, 16, 17 and 18 DNA constructs were based on the G protein binding domain, or RH domain, sequences described by the Structural Genomics Consortium (Soundararajan, Willard et al. 2008). RGS4 utilized rat DNA corresponding to RH domain residues 51-179. Human RGS5 and RGS8 constructs corresponding to RH domain residues 53-181 and 45-180, respectively, were employed. Gαi1, Gαq, RGS6Lα2, and Gβ5 DNA used coded for full-length human proteins.
RGS4-RH, RGS17-RH, RGS6Lα2, Gβ5, Gαi1 and Gαq DNA was cloned in to each of the four NanoBit vectors as described in manufacturers protocols. Restriction sites utilized were as follows: XhoI and NheI for RGS4, RGS6Lα2, Gβ5, and Gαq; EcoRI and Bgl II for RGS17; and XhoI and SacI for Gαi1. The remaining RGS-RH DNA was cloned in to vectors pBiT1.1-C and pBiT1.1-N (C and N terminal large bit fusions) only. Restriction sites utilized were as follows: NheI and SacI for RGS1; XhoI and SacI for RGS2, RGS10, and RGS14; XhoI and NheI for RGS5, RGS6, RGS7, RGS8, RGS16, RGS18, and cysteine-less RGS4 (C71A, C95A, C132A, C148A). Proper plasmid construction was confirmed by Sanger Sequencing (Iowa Institute of Human Genetics, University of Iowa). For clarity, the constructs will be referred to as follows. Fusion constructs with the NanoBit protein fragments on the N-terminus of the protein of interest will be represented as ‘LB-protein of interest’ or ‘SB-protein of interest’ for large and small bit respectively. Constructs with the bit pieces on the C-terminus of the protein of interest will be represented as ‘protein of interest-LB’ or ‘protein of interest-SB’ for large and small bit respectively (Figure 1).
Assessment of Vector Combinations for Protein Interaction Pairs
Prior to plating, 96-well half area clear bottom white plates (Corning 3885) were coated with poly-D-lysine and allowed to dry overnight. HEK293T cells were then plated at 20,000 cells per well and allowed to adhere overnight at 37°C in 5% CO2. Cells were then transfected using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA) per manufacturer protocols.
40-44 h post-transfection, media was aspirated and replaced with 40 µl Hank’s Balanced Salt Solution (HBSS) (Thermo Fisher Scientific, Waltham, MA) supplemented with HEPES pH 7.2-7.5 (Life Technologies, Carlsbad, CA) at a final concentration of 20 mM. A 5X stock of NanoGlo Live Cell Reagent was prepared per manufacture protocols in a reduced light environment and added to the wells of the assay plate at 10 µl per well. The assay plate was then read on a Synergy 2 plate reader (BioTek, Winooski, VT) in luminescence mode.
For RGS: Gα interactions, the first 30 min of read time were in the absence of any stimulation, and represent the baseline read. After the baseline read, wells were either treated with an aluminum fluoride salt solution to achieve final concentrations of 6.67 mM NaF and 83.33 µM AlCl3 (hereafter referred to as AlF4) or HBSS (vehicle) and signal was monitored for the duration of the read. Data analysis was performed using GraphPad Prism 7. For RGS: Gα interactions, area under the curve (AUC) analysis was used to quantify the difference in luminescence signal between AlF4 treated and vehicle treated conditions from the point of AlF4 addition to the end of the kinetic read (60 min total). Non-linear fit analysis using the equation Y=Bottom + (Top-Bottom) / (1+10^((LogEC50-X)*HillSlope) with no constraints was used to determine the time to 50% signal post AlF4 addition, where the values for Top and Bottom refer to the plateaus of the curve.
Treatment with small molecule inhibitor 6383479
Plates and cells were treated as described above. The transfection pair used was LB-RGS4: SB-Gαi1 or SmBit-PRKACA: LgBit-PRKAR2A control vectors, with transfection preparation as described above. 30 min post AlF4 addition, wells were treated with either 6383479 (final concentration of 31.6 µM, 0.32% DMSO) or vehicle (HBSS, 0.32% DMSO) and the plate was monitored for an additional 60 min. Effect of compound treatment was calculated via AUC analysis from point of compound addition to end of read.
Z-factor Determination
Plates were prepared and cells plated as described above. 48 wells were transfected with the optimized DNA pairs, and 48 wells were transfected with the respective LB-fusion and negative control vector in a similar manner as described above. For RGS: Gα interaction pairs, a 10X stock of NanoGlo Reagent was prepared and added to wells of the assay plate at 10 µl per well resulting in a 2X final concentration. For the RGS6L: Gβ5 interaction pair, a 5X stock of NanoGlo Reagent was prepared and added to wells of the assay plate at 10 µl per well resulting in a 1X final concentration. Assay plates were then read on a Synergy 2 plate reader for the indicated time in luminescence mode. For RGS: Gα interactions, the first 15 min of a read were in the absence of any stimulation, and represent the baseline read. After the baseline read, wells were either treated with AlF4 or vehicle and signal was monitored for the duration of the read. Data analysis was performed using GraphPad Prism 7.
Results
Determination of Optimized Protein Complementation Pairs for RGS4 and RGS17 with Gαi1
Appropriate combinations of RGS, Gα, and control vectors were co-transfected into HEK293T cells as described above and the interaction monitored over a period of 90 min (Figure 2A). Addition of AlF4 promotes formation of Gα: GDP: AlF4 complex, which mimics the Gα transition state and promote the high affinity RGS: Gα interaction (Kleuss, Raw et al. 1994, Berman, Kozasa et al. 1996, Tesmer, Berman et al. 1997). Not surprisingly, addition of AlF4 resulted in a robust signal increase over vehicle treated wells for all conditions tested (Figure S1). All interaction pairs were tested for the first two trials, with only the optimal four conditions for each RGS being tested for the third trial. Determination of optimal conditions was based on the net signal increase and the fold signal amplification in response to AlF4 addition (Figure S1).
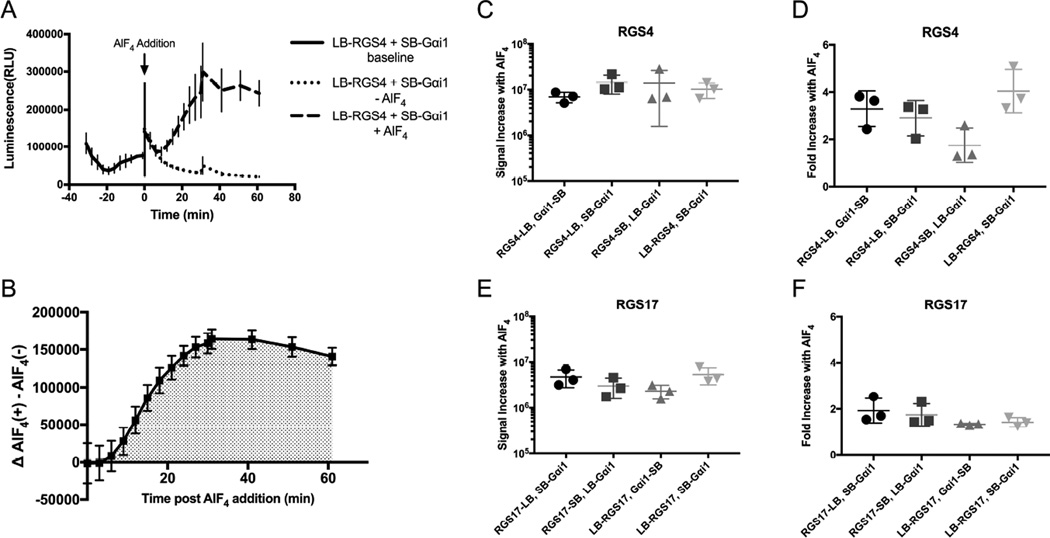
Figure 2.
Optimization of Vector Pairs Using RGS4 and RGS17. RGS:Gαi1 complementation pairs were optimized using RGS4 and RGS17. A.) Representative kinetic read of interaction of RGS4: Gαi1. B.) Delineation of time range for area under the curve analysis, starting at AlF4 addition. 2C-D is a comparison of four vector pairs for the RGS4: Gαi1 interaction shown as increase in area under the curve following AlF4 addition (C) and fold increase in area under the curve (D) upon stimulation with AlF4. The same analysis was performed for the four optimal RGS17: Gαi1 vector pairs (E/F). Data are n=3 in at least duplicate, ± SD.
The four optimal pairs for each were assessed via AUC analysis (Figures 2B-2F). The combinations of LB-RGS: SB-Gαi1, RGS-SB: LB-Gαi1, and RGS-LB: SB-Gαi1 were identified as three of the four optimal combinations for both RGS4 and RGS17. For RGS4, the four optimal vector pairs included the three listed above and RGS4-LB: Gαi1-SB. These resulted in a similar net signal increase in response to AlF4 treatment (Figure 2C), however one vector pair (LB-RGS4, SB-Gαi1) demonstrated greater fold signal amplification over vehicle treated control (Figures 2D). This vector pair was determined to be the optimal pair for RGS4. The vector pair LB-RGS17, SB-Gαi1 resulted in the greatest response to AlF4, and comparable signal amplification as the other vector conditions (Figure 2E, 2F). Since identical vector conditions (LB-RGS, SB-Gαi1 protein) resulted in the best responses for both RGS4 and RGS17, it was decided to move forward using this vector pairing.
Characterization of RGS: Gαi1 Interactions
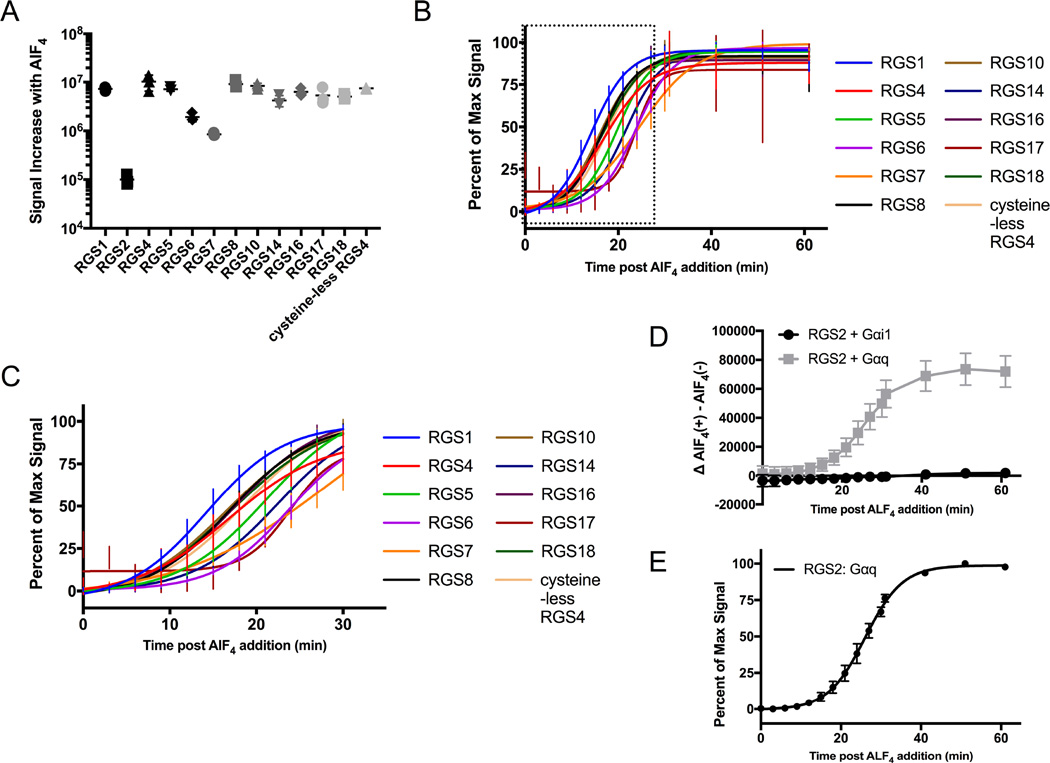
As protein interaction analysis of both RGS4 and RGS17 with Gαi1 identified the same vector combination as the optimized pair, a panel of 11 additional RGS RH domains was interrogated using this same vector pairing, where the RGS is tagged with LB on the N terminus. 10 of the 11 protein interaction pairs exhibited a response to addition of AlF4, with the only RGS not presenting a response being RGS2, which was expected as RGS2 is known to only interact with Gαq (Figure S2) (Soundararajan, Willard et al. 2008).
Analysis of the signal increase in response to AlF4 addition allowed for separation of response in to two distinct groups (Figure 3A). RGS1, RGS4, RGS5, RGS8, RGS10, RGS14, RGS16, RGS17, RGS18 and cysteine-less RGS4 produced the greatest net signal increase over vehicle treated control, while RGS6 and RGS7 produced a minimal response. The apparent response for RGS2 is a consequence of variation in low signal conditions, where the average signal for AlF4 treatment happens to rise above baseline. Examination of the response curves (Figure S2) demonstrates that the AlF4 and vehicle treated conditions for RGS2 are highly overlapping, such that no significant interaction of RGS2 with Gαi1 is detectable. Assessing the rate of formation of the RGS: Gα interaction in response to AlF4 by calculation of time elapsed to 50% response (Figure 3B, 3C, Table 1) allows for the binning of response into two groups, where the slowest rate of formation in the more rapid response group has no overlap with the fastest rate of formation in the gradual response group at the 95% CI. RGS1, RGS4, RGS5, RGS8, RGS10, RGS16, RGS18, and cysteine-less RGS4 all demonstrate the most rapid rate of RGS: Gα interaction formation in response to AlF4 stimulus. RGS6, RGS7, RGS14, and RGS17 exhibit a slower rate of protein interaction formation. Analysis of the two response groups indicates that the rate of protein interaction formation is, to a degree, independent of the magnitude of response. For example, RGS17 exhibits a net signal increase in response to AlF4 that is 2.8 and 6.3 fold greater than RGS6 and RGS7 respectively, yet has an overlapping rate of RGS: Gα interaction formation with these two RGSs.
Figure 3.
Characterization of 12 WT and 1 Mutant RGS Proteins Interaction with Gαi1 or Gαq. A.) Depicts the net increase in area under the curve in response to AlF4 stimulation for 13 RGS: Gαi1 protein pairs. B.) Normalized non- linear fit of luminescence over time to assess the rate of protein interaction. C.) Magnification of the boxed area in panel B. D.) RGS2 interaction with Gαi1 and Gαq in response to AlF4. E.) Non-linear fit of RGS2: Gαq as in panel B. Data are n=3 in at least duplicate, ± SD. Data for panel D are the background subtracted results of n=3 in at least duplicate, ± SEM.
Table 1.
Rate of Interaction Formation. Rates are the time to 50% signal calculated from curves in Figure 3B and 3E. Calculated values shown as 95% CI.
| RGS | Time to 50% Response (min) |
|---|---|
| RGS1 | 13.2 to 14.9 |
| RGS2 (Gαq) | 25.6 to 26.5 |
| RGS4 | 14.7 to 18.8 |
| RGS5 | 18.7 to 20.7 |
| RGS6 | 23.2 to 24.4 |
| RGS7 | 23.3 to 25.7 |
| RGS8 | 15.3 to 17.4 |
| RGS10 | 14.8 to 16.9 |
| RGS14 | 20.8 to 22.7 |
| RGS16 | 15.2 to 17.5 |
| RGS17 | 22.0 to 25.5 |
| RGS18 | 15.6 to 18.0 |
| Cysless RGS4 | 16.1 to 18.5 |
Detecting RGS2: Gαq Interaction
The R4 RGS protein RGS2 is unique in that it only interacts with Gαq (Heximer, Watson et al. 1997). Since the ability to test small molecules for inhibition of this RGS would advantageous, interaction of RGS2 and Gαq was investigated using the NanoBit system. A robust increase in signal was detected for the RGS2: Gαq interaction when cells were treated with AlF4 (Figure 3D). The rate of formation of the RGS2: Gαq interaction was investigated in the same manner as RGS: Gαi1 interactions. Although these interactions use different Gα protein partners, the RGS2: Gαq interaction forms at the slowest rate of any of the protein interactions tested (Figure 3E, Table 1).
RGS4: Gαi1 Interaction Disrupted by Compound 6383479
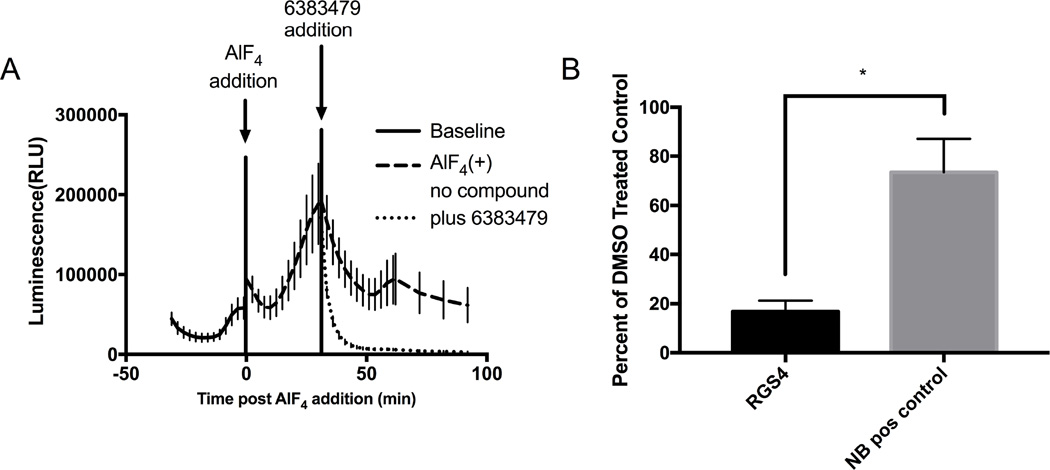
Having shown that the interaction between RGS proteins and Gαi1 is detectable using this system, we tested if the interaction could be disrupted with a known RGS4 inhibitor, 6383479 (Storaska, Mei et al. 2013) (Figure 4A). Addition of 6383479 resulted in an 83% decrease in luminescence signal compared to vehicle as calculated by AUC. To ensure this was due to disruption of the RGS4: Gαi1 interaction and not due to inhibition of the assay, the constitutive positive control pair of the catalytic and regulatory domains of PKA (PRKACA and PRKAR2A respectively) was tested with 6383479. This pair retained 73% of vehicle treated signal, indicating reduction observed for RGS4: Gαi1 is specific for the RGS: Gα pair and not due to assay interference. (Figure 4B). This demonstrates that the identification of potential RGS inhibitors via disruption of the RGS and Gα interaction is observable using this system.
Figure 4.
Selective Disruption of RGS4: Gαi1 Interaction with 6383479. Small molecule inhibition of RGS4: Gαi1 interaction. A) n=3 raw luminescence trace of RGS4: Gαi1 in the presence or absence of RGS4 inhibitor 6383479. B) Quantification of 6383479 effect on RGS4: Gαi1 and control using AUC-compound treated/AUC-vehicle treated. Data are n=3 in at least duplicate, ± SD. P-value = 0.012
Observation of the RGS6Lα2: Gβ5 Interaction
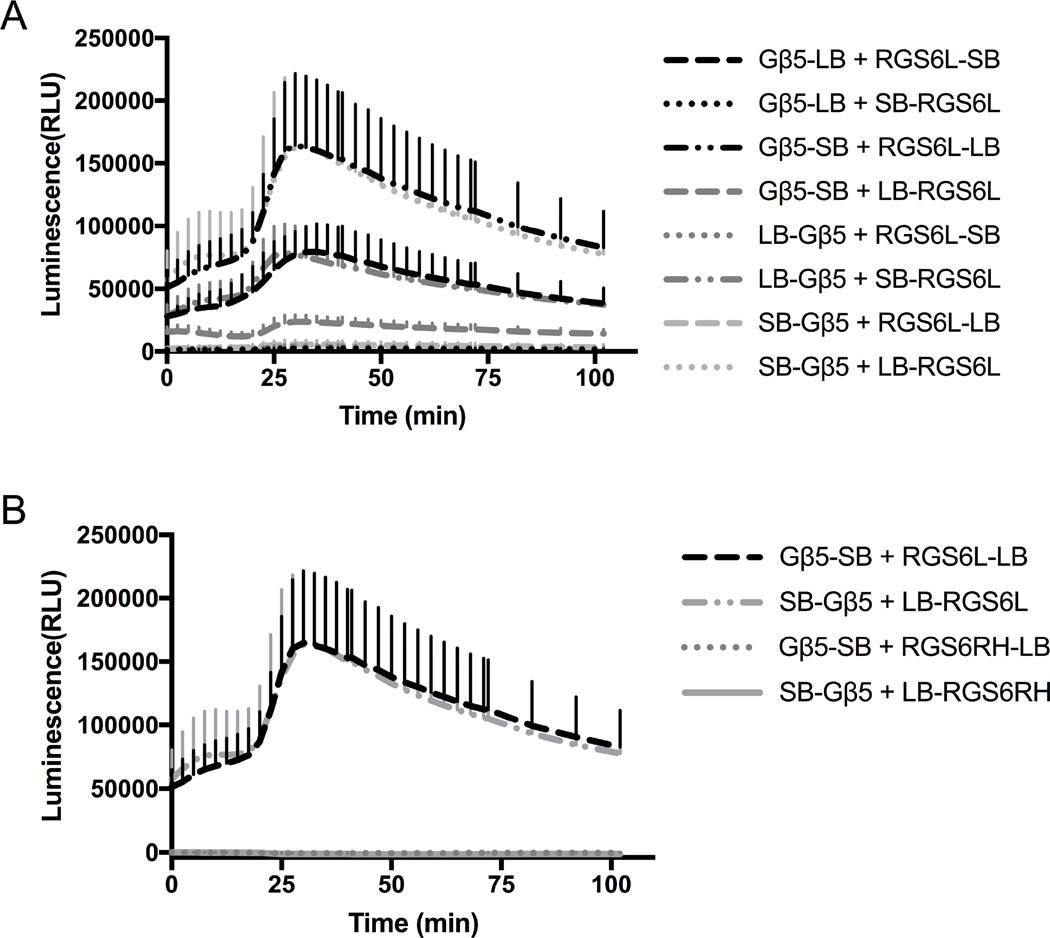
RGS6 is capable of interacting with, and therefore eliciting signaling through proteins other than Gα subunits (Liu, Chatterjee et al. 2002, Liu and Fisher 2004, Maity, Yang et al. 2011). Here, we analyzed the RGS6Lα2: Gβ5 interaction using the NanoBit system (Figure 5A). Only combinations where both bit pieces were on either the N or the C termini of both proteins produced a luminescence signal. The interactions producing the greatest signal consisted of large bit-RGS6L fusions and small bit-Gβ5 fusions (Gβ5-SB: RGS6L-LB and SB-Gβ5: LB-RGS6L). To prove that the interaction was due to one of the accessory domains and not due to an interaction with the RH domain of RGS6Lα2, we tested the combinations of LB-RGS6RH: SB-Gβ5 and RGS6RH-LB: Gβ5-SB (Figure 5B). As expected, these combinations did not produce a luminescence signal above baseline.
Figure 5.
Characterization of RGS6L:Gβ5 Interaction. Interaction of RGS6L with Gβ5 was also detectable using this system. A.) Raw traces of RGS6Lα2: Gβ5 vector pairs. B.) Raw trace comparison of RGS6Lα2: Gβ5 interactions producing largest signal and the corresponding RGS6RH: Gβ5 pairing. Data are n=3 in at least duplicate, ± SD.
Suitability of NanoBit Platform for RGS Screening Assays
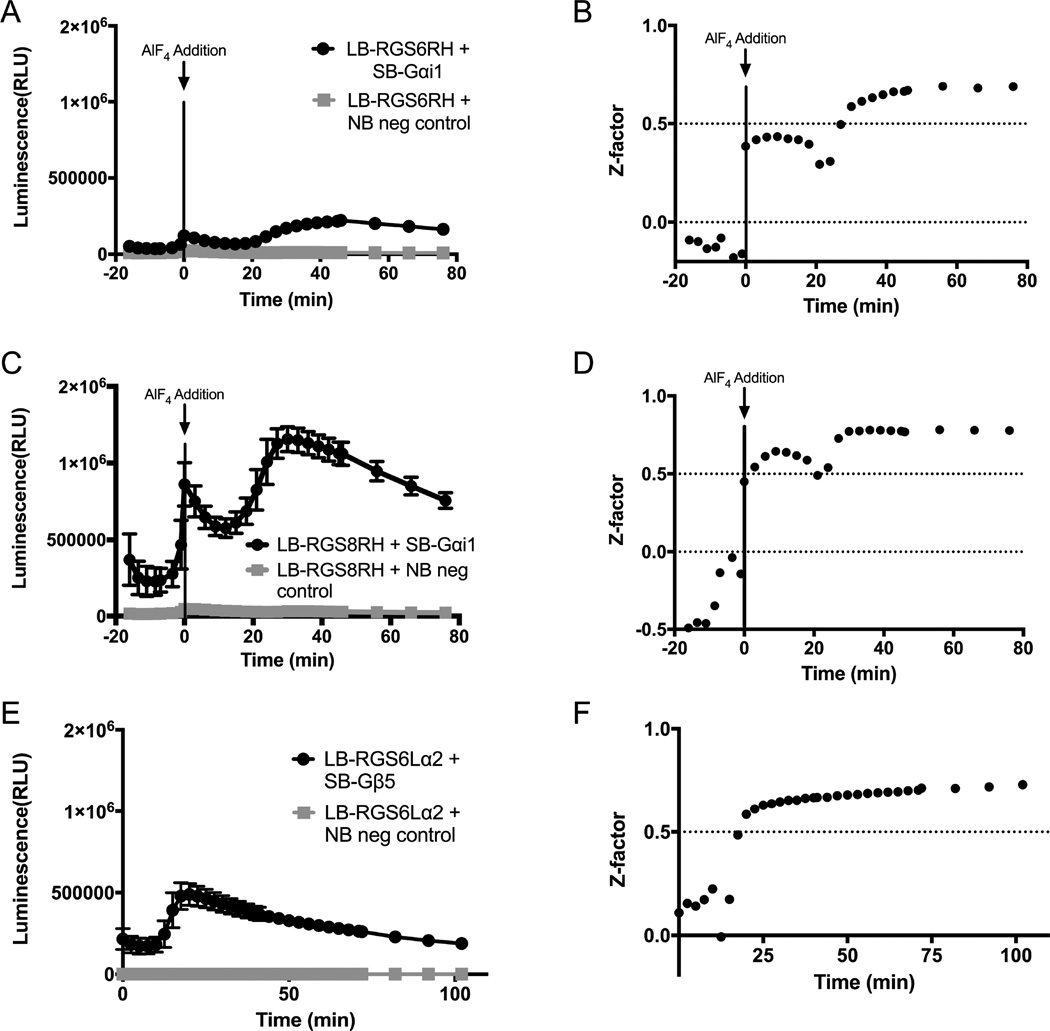
Having demonstrated the use of this protein complementation assay allowed for the characterization of RGS: Gα interactions as well as the assessment of the RGS6L: Gβ5 interaction, we assessed the amenability of the system to high throughput screening, determined via calculation of a Z-factor given by the equation:
Where σp and σn represent the standard deviations of the positive and negative controls respectively and µp and µn represent the averages of the positive and negative controls respectively (Zhang, Chung et al. 1999). 96 well full plate Z-factors were calculated for RGS6RH: Gαi1 (Figure 6A-B), RGS8RH: Gαi1 (Figure 6C-D), and RGS6Lα2: Gβ5 (Figure 6E-F) protein-protein interactions. Since the NanoBit system allows for a kinetic measurement, the robustness of the assay over time was assessed. Initial experiments with RGS6RH and RGS8RH and 1X final NanoGlo Live Cell Reagent did not produce a Z-factor that is considered acceptable for a high throughput screening campaign, with Z-factors of 0.39 and 0.44, respectively (data not shown). However, increasing the final NanoGlo Reagent concentration to 2X produced acceptable Z-factors due to an increased difference between the positive and negative control average values (i.e. µp and µn). Production of luminescence signal is a product of enzymatic consumption of the substrate (furimazine), and the increased luminescence signal using 2X reagent is likely due to a corresponding increase in substrate availability. For RGS6RH, the Z-factor crosses the threshold for an acceptable high throughput assay of 0.5 at 30 min post AlF4 addition, reaching a maximum of 0.69 at 56 min post AlF4 addition (Figure 6B). Once the assay reaches an acceptable threshold at 30 min, it remains stable for the duration of the read, an additional 46 min. For RGS8RH, the Z-factor becomes stable above 0.5 starting at 24 min post AlF4 addition and remains stable for the duration of the read, an additional 52 min (Figure 6D). The dip in Z-factor shown at 20 min corresponds to the low point in signal in the kinetic read just before the system starts to demonstrate response to the addition of AlF4 (Figure 6C). For the RGS6L: Gβ5 interaction, a 1X final concentration of NanoGlo Reagent is sufficient to produce an acceptable Z-factor. The Z-factor crosses the acceptable threshold at 20 min post addition of NanoGlo Reagent and continues to increase for the duration of the read, reaching a maximum of 0.73 at 102 min post addition of reagent (Figure 5F).
Figure 6.
Amenability of RGS NanoBit™ Systems to High Throughput Screening. Suitability of NanoBit™ systems for high throughput campaigns. A/B.) The low magnitude interaction of RGS6RH: Gαi1 in a 96 well half area plate. The raw signal over time (A) along with Z-factor over time (B) are shown. C/D.) The high magnitude interaction of RGS8RH: Gαi1 was also tested in a similar manner. E/F.) Finally the RGS6L: Gβ5 interaction was assessed. Y-axis normalized to RGS8RH: Gαi1 signal to demonstrate varying magnitude of signal between protein pairs assessed.
Discussion
We have implemented and characterized an in-cell protein interaction assay that is amenable to both primary discovery and selectivity profiling for RGS inhibitors. As part of this study, we have also established that our characterization of RGS RH domain interactions with AlF4 activated Gαi1 and Gαq is in agreement with the prior published biochemical binding profiles of these proteins (Heximer, Watson et al. 1997, Hooks, Waldo et al. 2003, Soundararajan, Willard et al. 2008). Our data shows that this protein complementation system is suitable for the investigation of RGS: Gα protein pairs.
In addition to establishing this technique for RGS protein interactions, our data also provide some insight into the relative affinity of different RGS proteins for Gα subunits. Analyzing the net increase of signal in response to AlF4 stimulation revealed information about relative interaction affinity among the RGS: Gα pairs. Additionally, analysis of the rate of signal amplification due to AlF4 stimulation demonstrated discrete differences in the rate of formation of the protein-protein interactions. This may give clues to how signals are regulated by RGS proteins in the cell. For example, a rapid and robust formation of an RGS: Gα interaction may suggest that RGS is essential for regulation of acute signaling events, while a gradual and moderate formation of an RGS: Gα interaction may suggest regulation of sustained signaling events. Further exploration of these phenomena may provide additional insight into RGS regulation of signaling events.
The NanoBit system is amenable to the assessment of a panel of RGS proteins with both Gαi1 and, for RGS2, Gαq. Therefore, this system is a powerful tool in the interrogation of small molecules for selectivity amongst RGS proteins in a cellular context. Most importantly in this regard, this system allows for the robust assessment of multiple RGS proteins using identical cellular conditions. In addition, the amenability of the system to quickly express and test point mutants, such as the exemplar cysteine-less RGS4 mutant presented here, shows this system can be used to assess small molecule mechanism of action in a cellular context as well. For example, a small molecule that also disrupts the interaction of the cysteine-less RGS4 mutant may indicate a cysteine independent mechanism of action, and further investigations utilizing point mutants to test for potential small molecule interactions with RGS proteins could be performed far more efficiently than purifying proteins representing individual point mutants.
Validation for high throughput screening was demonstrated by several lines of experimentation. First, the specific reduction of luminescence signal upon treatment with a known RGS4 inhibitor shows that this system may be used for the identification of novel small molecules through high throughput screening efforts. Second, this assay is amenable to a high throughput campaign as determined by a Z-factor calculation. The only other published method used for the observation of disruption of an RGS: Gα interaction in cells is the GFP-RGS fusion method, which requires the use of confocal microscopy and significant post hoc analysis. In comparing NanoBit to the calcium flux high throughput paradigm, this system allows for the assessment of small molecule activity at the protein interaction level. While a functional assay may still be preferable, small molecules that elicit a change in functional interrogation systems may in fact be affecting a downstream target rather than the RGS, and may require extensive follow-up experimentation to determine mechanism of action.
The result that both RGS6RH and RGS8RH were able to produce acceptable Z-factors suggests that, at least for the Gαi1 system, other RGS: Gα pairs may be amenable to a high throughput campaign, as RGS6RH produced a low net response while RGS8RH had one of the greatest. The result that RGS6Lα2: Gβ5 yielded an acceptable Z-factor is encouraging because it opens the possibility of targeting RGS mediated signaling that does not include targeting the RGS: Gα protein-protein interaction. Interaction with Gβ5 is a characteristic unique to the R7 family of RGS proteins and small molecule disrupting this interaction would not be expected to interrupt non-R7 family RGS function, which would increase the selectivity of the molecule and reduce potential off target consequences of a pan RGS inhibitor.
In conclusion, we characterized the interactions of a panel of RGS proteins with Gαi1 and of RGS2 with Gαq as well as the interaction of RGS6Lα2 with its obligatory binding partner Gβ5 using this Protein Complementation Assay. This is the largest implementation of this system to date, characterizing 15 separate protein-protein interactions. The platform was deemed suitable for a high throughput screening campaign via Z-factor calculation, and the demonstration that protein pair disruption via small molecules is observable lends validity to the use of the platform for drug discovery efforts. In addition, the system provided interesting kinetic data with regard to relative RGS: G protein complex formation among different RGS proteins. Finally, the inclusion and validation of mutant RGS proteins indicates that this platform will not only be integral in future cell based discovery efforts, but also capable of mechanistic interrogation of compound binding site and function in a cellular context.
Supplementary Material
Characterization of 14 RGS: Gα interactions using NanoBit
RGS4: Gαi1 interaction disruption with compound 6383479
Characterization of RGS6Lα2: Gβ5 interaction using NanoBit
Calculation of Z-factors for RGS6RH: Gαi1, RGS8RH: Gαi1, and RGS6Lα2: Gβ5
Acknowledgments
We would like to thank Zachariah Builta for his role in the construction of RGS4 and RGS17 plasmids.
Funding: This work was supported by NIH 5R01CA160470 (DLR), NIH T32GM067795 (MPH) and American Foundation for Pharmaceutical Education Predoctoral Fellowship (CRB and MPH)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berman DM, Kozasa T, Gilman AG. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem. 1996;271(44):27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- Blazer LL, Storaska AJ, Jutkiewicz EM, Turner EM, Calcagno M, Wade SM, Wang Q, Huang XP, Traynor JR, Husbands SM, Morari M, Neubig RR. Selectivity and anti-Parkinson's potential of thiadiazolidinone RGS4 inhibitors. ACS Chem Neurosci. 2015;6(6):911–919. doi: 10.1021/acschemneuro.5b00063. [DOI] [PubMed] [Google Scholar]
- Blazer LL, Zhang H, Casey EM, Husbands SM, Neubig RR. A nanomolar-potency small molecule inhibitor of regulator of G-protein signalling proteins. Biochemistry. 2011;50(15):3181–3192. doi: 10.1021/bi1019622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosier B, Doyen PJ, Brolet A, Muccioli GG, Ahmed E, Desmet N, Hermans E, Deumens R. Inhibition of the regulator of G protein signalling RGS4 in the spinal cord decreases neuropathic hyperalgesia and restores cannabinoid CB1 receptor signalling. Br J Pharmacol. 2015;172(22):5333–5346. doi: 10.1111/bph.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc Natl Acad Sci U S A. 2003;100(11):6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP, Wood KV. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem Biol. 2016;11(2):400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- Garnier M, Zaratin PF, Ficalora G, Valente M, Fontanella L, Rhee MH, Blumer KJ, Scheideler MA. Up-regulation of regulator of G protein signaling 4 expression in a model of neuropathic pain and insensitivity to morphine. J Pharmacol Exp Ther. 2003;304(3):1299–1306. doi: 10.1124/jpet.102.043471. [DOI] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7(11):1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MP, Roman DL. Regulator of G Protein Signaling 17 as a Negative Modulator of GPCR Signaling in Multiple Human Cancers. AAPS J. 2016;18(3):550–559. doi: 10.1208/s12248-016-9894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heximer SP, Watson N, Linder ME, Blumer KJ, Hepler JR. RGS2/G0S8 is a selective inhibitor of Gqalpha function. Proc Natl Acad Sci U S A. 1997;94(26):14389–14393. doi: 10.1073/pnas.94.26.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks SB, Waldo GL, Corbitt J, Bodor ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J Biol Chem. 2003;278(12):10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 2009;78(10):1289–1297. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- James MA, Lu Y, Liu Y, Vikis HG, You M. RGS17, an overexpressed gene in human lung and prostate cancer, induces tumor cell proliferation through the cyclic AMP-PKA-CREB pathway. Cancer Res. 2009;69(5):2108–2116. doi: 10.1158/0008-5472.CAN-08-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleuss C, Raw AS, Lee E, Sprang SR, Gilman AG. Mechanism of GTP hydrolysis by G-protein alpha subunits. Proc Natl Acad Sci U S A. 1994;91(21):9828–9831. doi: 10.1073/pnas.91.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Kreitzer AC. RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron. 2012;73(2):347–359. doi: 10.1016/j.neuron.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li L, Lin J, Hu X, Li B, Xue A, Shen Y, Jiang J, Zhang M, Xie J, Zhao Z. Deregulation of RGS17 Expression Promotes Breast Cancer Progression. J Cancer. 2015;6(8):767–775. doi: 10.7150/jca.11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chatterjee TK, Fisher RA. RGS6 interacts with SCG10 and promotes neuronal differentiation. Role of the G gamma subunit-like (GGL) domain of RGS6. J Biol Chem. 2002;277(40):37832–37839. doi: 10.1074/jbc.M205908200. [DOI] [PubMed] [Google Scholar]
- Liu Z, Fisher RA. RGS6 interacts with DMAP1 and DNMT1 and inhibits DMAP1 transcriptional repressor activity. J Biol Chem. 2004;279(14):14120–14128. doi: 10.1074/jbc.M309547200. [DOI] [PubMed] [Google Scholar]
- Mackie DI, Roman DL. Development of a novel high-throughput screen and identification of small-molecule inhibitors of the Galpha-RGS17 protein-protein interaction using AlphaScreen. J Biomol Screen. 2011;16(8):869–877. doi: 10.1177/1087057111410427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity B, Yang J, Huang J, Askeland RW, Bera S, Fisher RA. Regulator of G protein signaling 6 (RGS6) induces apoptosis via a mitochondrial-dependent pathway not involving its GTPase-activating protein activity. J Biol Chem. 2011;286(2):1409–1419. doi: 10.1074/jbc.M110.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy CA, Mackie DI, Roman DL. A high throughput screen for RGS proteins using steady state monitoring of free phosphate formation. PLoS One. 2013;8(4):e62247. doi: 10.1371/journal.pone.0062247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Ota S, Neubig RR. Polyplexed flow cytometry protein interaction assay: a novel high-throughput screening paradigm for RGS protein inhibitors. J Biomol Screen. 2009;14(6):610–619. doi: 10.1177/1087057109336590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71(1):169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- Roof RA, Roman DL, Clements ST, Sobczyk-Kojiro K, Blazer LL, Ota S, Mosberg HI, Neubig RR. A covalent peptide inhibitor of RGS4 identified in a focused one-bead, one compound library screen. BMC Pharmacol. 2009;9:9. doi: 10.1186/1471-2210-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren B. Regulator of G protein signaling proteins as drug targets: current state and future possibilities. Adv Pharmacol. 2011;62:315–347. doi: 10.1016/B978-0-12-385952-5.00002-6. [DOI] [PubMed] [Google Scholar]
- Snow BE, Betts L, Mangion J, Sondek J, Siderovski DP. Fidelity of G protein beta-subunit association by the G protein gamma-subunit-like domains of RGS6, RGS7, and RGS11. Proc Natl Acad Sci U S A. 1999;96(11):6489–6494. doi: 10.1073/pnas.96.11.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Krumins AM, Brothers GM, Lee SF, Wall MA, Chung S, Mangion J, Arya S, Gilman AG, Siderovski DP. A G protein gamma subunit-like domain shared between RGS11 and other RGS proteins specifies binding to Gbeta5 subunits. Proc Natl Acad Sci U S A. 1998;95(22):13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov E, Iannitti DA, Schrum LW, McKillop IH. Altered expression and function of regulator of G-protein signaling-17 (RGS17) in hepatocellular carcinoma. Cell Signal. 2011;23(10):1603–1610. doi: 10.1016/j.cellsig.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundstrom M, Doyle DA, Siderovski DP. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits. Proc Natl Acad Sci U S A. 2008;105(17):6457–6462. doi: 10.1073/pnas.0801508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Maity B, Anderegg SP, Allamargot C, Yang J, Fisher RA. Regulator of G protein signaling 6 is a critical mediator of both reward-related behavioral and pathological responses to alcohol. Proc Natl Acad Sci U S A. 2015;112(7):E786–E795. doi: 10.1073/pnas.1418795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Maity B, Fisher RA. Two for the Price of One: G Protein-Dependent and -Independent Functions of RGS6 In Vivo. Prog Mol Biol Transl Sci. 2015;133:123–151. doi: 10.1016/bs.pmbts.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Stewart A, Maity B, Wunsch AM, Meng F, Wu Q, Wemmie JA, Fisher RA. Regulator of G-protein signaling 6 (RGS6) promotes anxiety and depression by attenuating serotonin-mediated activation of the 5-HT(1A) receptor-adenylyl cyclase axis. FASEB J. 2014;28(4):1735–1744. doi: 10.1096/fj.13-235648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storaska AJ, Mei JP, Wu M, Li M, Wade SM, Blazer LL, Sjogren B, Hopkins CR, Lindsley CW, Lin Z, Babcock JJ, McManus OB, Neubig RR. Reversible inhibitors of regulators of G-protein signaling identified in a high-throughput cell-based calcium signaling assay. Cell Signal. 2013;25(12):2848–2855. doi: 10.1016/j.cellsig.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89(2):251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- Vashisth H, Storaska AJ, Neubig RR, Brooks CL., 3rd Conformational dynamics of a regulator of G-protein signaling protein reveals a mechanism of allosteric inhibition by a small molecule. ACS Chem Biol. 2013;8(12):2778–2784. doi: 10.1021/cb400568g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisler JW, Xiao K, Thomsen AR, Lefkowitz RJ. Recent developments in biased agonism. Curr Opin Cell Biol. 2014;27:18–24. doi: 10.1016/j.ceb.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Maity B, Huang J, Gao Z, Stewart A, Weiss RM, Anderson ME, Fisher RA. G-protein inactivator RGS6 mediates myocardial cell apoptosis and cardiomyopathy caused by doxorubicin. Cancer Res. 2013;73(6):1662–1667. doi: 10.1158/0008-5472.CAN-12-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Woo J, Park JO, Choi EJ, Shin HS, Roh DH, Kim KS. Intrathecal RGS4 inhibitor, CCG50014, reduces nociceptive responses and enhances opioid-mediated analgesic effects in the mouse formalin test. Anesth Analg. 2015;120(3):671–677. doi: 10.1213/ANE.0000000000000607. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.