Figure 5.
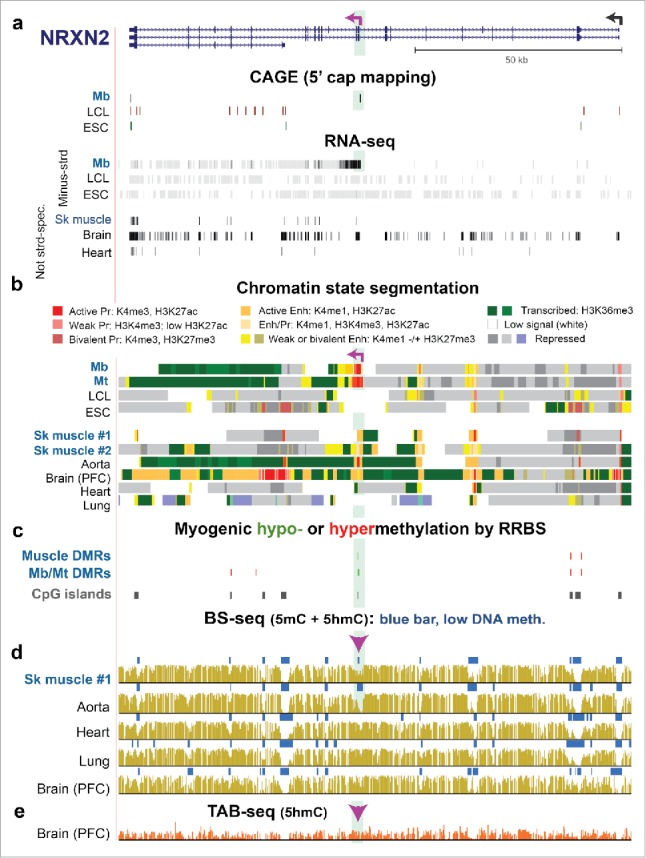
NRXN2, a neuronal gene, displays a Mb/Mt-specific alternative promoter whose DNA hypomethylation persists in SkM despite the loss of promoter activity. (a) RefSeq gene isoforms structures for NRXN2 (chr11:64,371,048–64,493,639) and RNA-seq as in Fig. 3 but also with the ENCODE profile of 5′ cap mapping (CAGE).37 Purple broken arrow on left, TSS for the Mb-associated transcript. (b) Chromatin state segmentation. (c) Significant hyper- or hypomethylated DMRs from 33 RRBS profiles.24 (d) and (e) Bisulfite-seq and TAB-seq. Highlighted green region, Mb/Mt-specific promoter region within NRXN2. Arrowhead, Epimark-tested site with high 5hmC in cerebellum (Fig. 2a).
