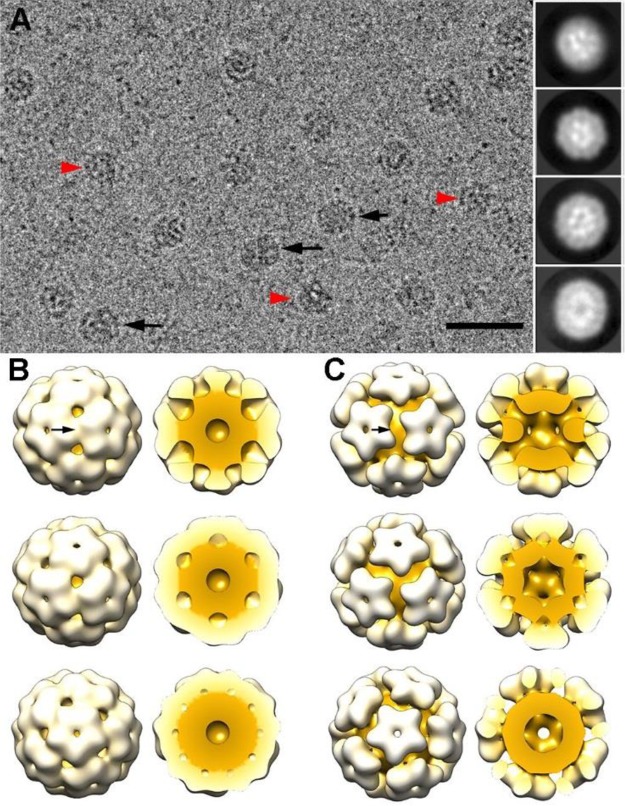
Figure 4.
Three-dimensional cryo-EM reconstructions of GOx–ssDNA-loaded CCMV capsids. (A) Cryo-electron micrograph of GOx–ssDNA-loaded CCMV capsids. Black arrows indicate elongated particles, and red arrowheads indicate irregular particles. Two-dimensional class averages derived from the final 15481 particle data set (inset). Bar, 50 nm. (B) Surface-shaded representation of the outer surface of the class I T = 1 capsid (diameter 21.4 nm) viewed along a 2-, 3-, and 5-fold axis of icosahedral symmetry (top to bottom). Models of the class I T = 1 capsid, with the front half of the cargo and protein shell removed (right). Protein shell is white, cargo is yellow. Arrow indicates a 2-fold axis of icosahedral symmetry. (C) Surface-shaded representations of the outer surface of the class II T = 1 capsid (diameter 22.6 nm) (as in B).