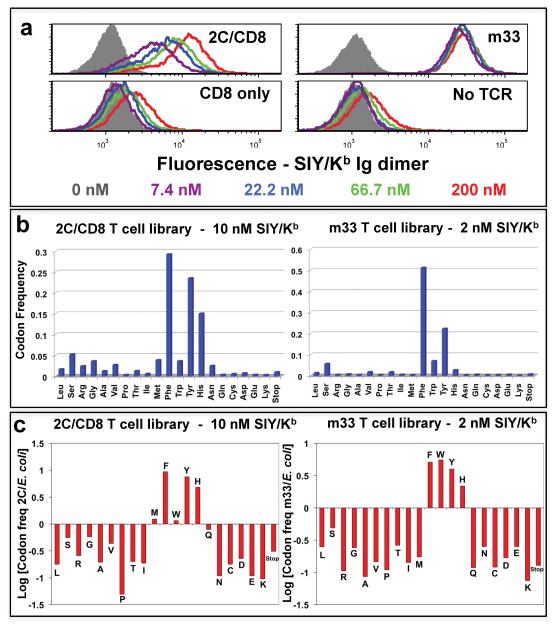
Figure 2. Selection of SIY/Kb-binding “signature” residues in TCRs at position 46β.
(a) SIY/Kb binding to T cell hybridoma cells transduced with 2C (left, top panel) or m33 (right, top panel) TCRs, with or without co-expression of CD8αβ, respectively. T cell hybridomas expressed with (top panels) or without (bottom panels) the TCRs were stained with the indicated concentrations of SIY/Kb Ig-dimer, followed by PE-labeled goat anti-mouse Ig, and analyzed by flow cytometry. (b) Frequency of residues from SIY/Kb-selected TCR libraries. T cell hybridomas were transduced with a library of TCRs with variation at position 46β in the 2C or m33 TCR, with or without co-expression of CD8αβ, respectively. The top 1.4% of SIY/Kb Ig dimer-binding T cells were sorted, RNA was isolated, and cDNA was used for PCR, followed by 454 sequencing. The frequencies measured for each amino acid residue at position 46β are shown for the 2C TCR library (selected with 10 nM SIY/Kb Ig-dimer, left panel, representing 29,754 sequences) and m33 TCR library (selected with 2 nM SIY/Kb Ig-dimer, right panel, representing18,560 sequences). (c) Relative representation of individual amino acid residues from the original, pre-selection library were analyzed by plotting the logarithm of the frequency of each residue in the SIY/Kb selected library divided by the frequency for that residue in the original E. coli library (Log[residue frequency, selected/residue frequency, original]). A value of +1 indicates the residue is 10-fold over-represented compared to the unselected library, while a value of −1 indicates the residue is 10-fold under-represented compared to the unselected library. Representative data are shown from four experiments. For the signature residues (Met, Phe, Trp, Tyr, and His) selected from the 2C library, the Tyr, Phe, and His residues were significantly enhanced (p<0.001). For the signature residues selected from the m33 library, the Tyr, Phe, His, and Trp residues were significantly enhanced, and the Met was significantly reduced (p<0.001)