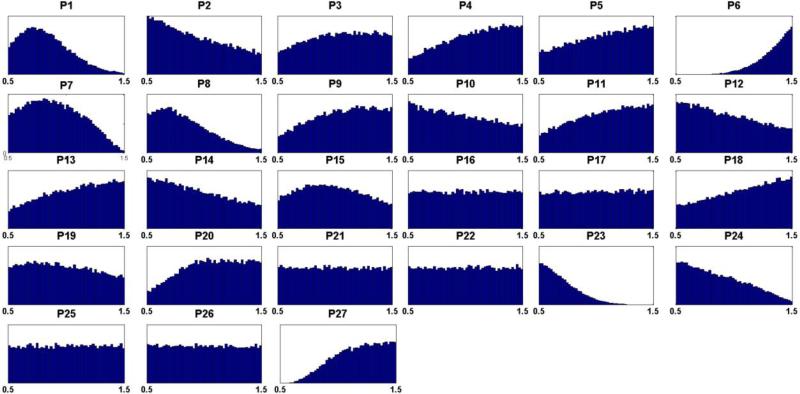
Figure 3. Distributions of hypothesized cancer actions from admissible sets for each candidate site.
Hypothesized cancer actions are identified by causing the same sign of change (either increased (+) or decreased (−)) in metabolites between normal cells and tumor represented by metabolomics data. Out of five million Monte Carlo simulations, a very small subpopulation (31,497 sets) remained after filtering according to the first, qualitative criterion and was retained in the form of admissible sets. X-axes are fold changes at each candidate site with respect to their nominal levels. The list (P1 – P27) is composed of: phosphoribosylpyrophosphate synthetase (P1), amidophosphoribosyltransferase (P2), hypoxanthine-guanine phosphoribosyltransferase (P3), adenine phosphoribosyltransferase (P4), ‘pyrimidine synthesis’ (P5), inosine monophosphate dehydrogenase (P6), guanosine monophosphate synthetase (P7), adenylosuccinate synthetase (P8), adenylosuccinate lyase (P9), guanosine monophosphate reductase (P10), adenosine monophosphate deaminase (P11), methionine adenosyltransferase (P12), protein O-methyltransferase (P13), s-adenosylmethionine decarboxylase (P14), 5'-Nucleotidase (P15), 5'(3') Nucleotidase (P16), diribonucleotide reductase (P17), adenosine deaminase (P18), RNA polymerase (P19), RNases (P20), DNA polymerase (P21), DNases (P22), xanthine oxidase/xanthine dehydrogenase (P23), guanine hydrolase (P24), ‘hypoxanthine excretion’ (P25), ‘xanthine excretion’ (P26), ‘uric acid excretion’ (P27).