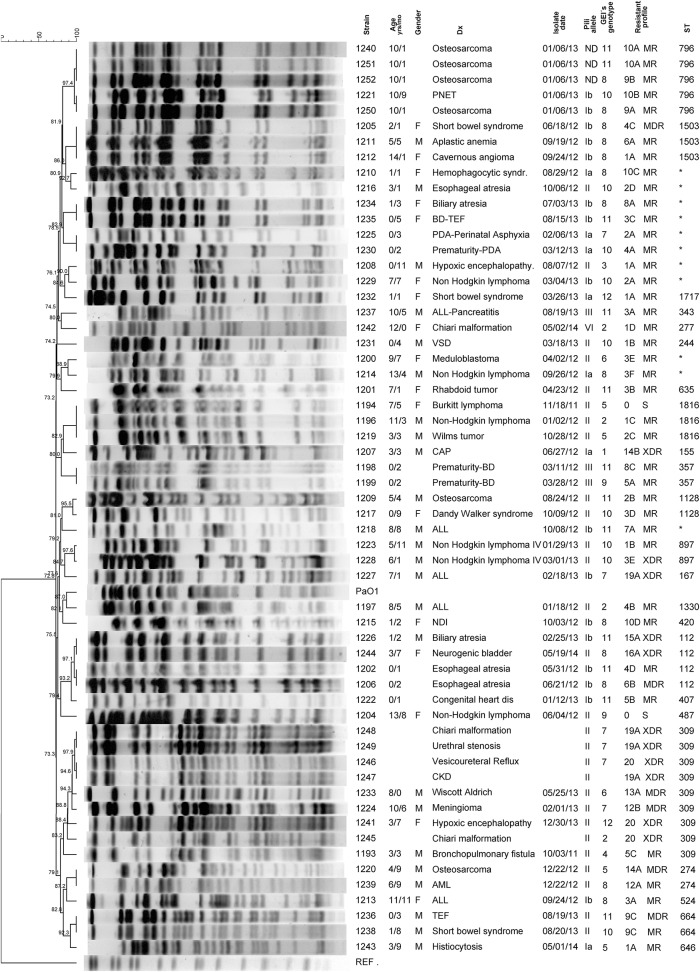
Figure 2.
Pulse-Field gel electrophoresis (PFGE) profile dendrogram and genetic and phenotypic characteristics of P. aeruginosa strains isolated from children with bacteremia. The dendrogram was generated by Dice similarity coefficient (Dice, 1945) and UPGMA (Day and Edelsbrunner, 1984) clustering methods by using PFGE images of SpeI digested genomic DNA. The scale bar shows the correlation coefficient (%). Underlying disease (Dx): PNET, primary neuroectodermal tumor; BD-TEF, bronchopulmonary dysplasia-tracheoesophageal fistula; PDA, persistent ductus arteriosus; SGER, severe gastroesophageal reflux; ALL, acute lymphoblastic leukemia; VSD, ventricular septal defect; BD, bronchopulmonary dysplasia; CAP, community acquired pneumonia; NDI, nephrogenic diabetes insipidus; CKD, chronic kidney disease; TEF, tracheoesophageal fistula; AML, acute myeloid leukemia. Asterisk indicate a new ST, which has not been assigned. MDR, Multi-Drug Resistant; MR Moderately Resistant; XDR Extensively drug Resistant. The pili alleles were obtained according to Kus's characterization (Kus et al., 2004), in which P. aeruginosa type IV pili are divided into five distinct phylogenetic groups. The GEIs genotype was assigned base on the presence/absence of genomic island, 12 different GEIs genotypes were found (for details see Table S2, Supplementary Material). The resistance profile was formed by a number and a letter: the number indicates how many antibiotics the strain was resistant to; the letters were assigned alphabetically to differentiate among the antimicrobial combinations for which the strains were resistant (detailed information is shown in Table S3, Supplementary Material).