Fig. 1.
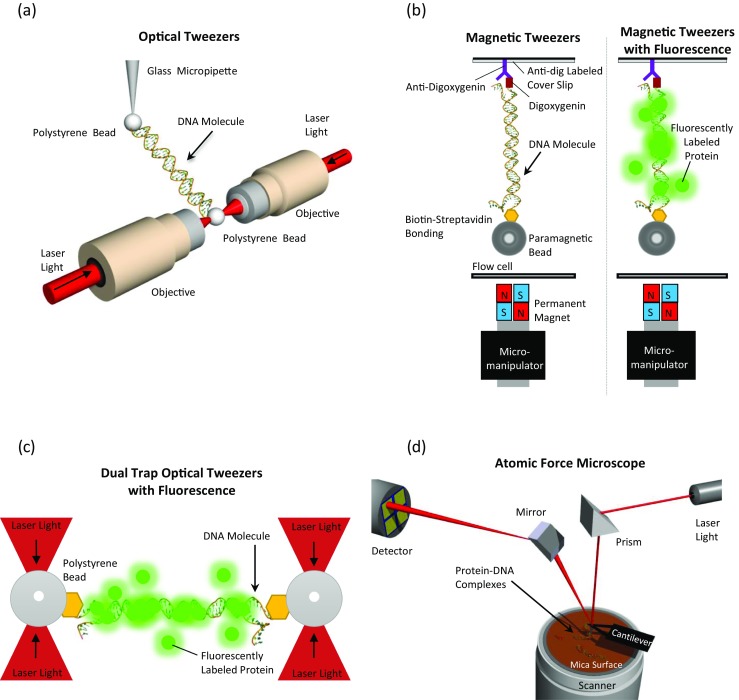
Schematic illustrations (not to scale) depicting single-molecule techniques used to investigate HMGB architectural protein binding to DNA. Optical tweezers, magnetic tweezers and atomic force microscopy are used. a In an optical tweezers setup, DNA tethered between labeled beads is extended and released. A glass micropipette tip is used to extend the DNA molecule, while on the other extremity, the deflection of the laser beam during extension is recorded and the signal is then translated into force. (From Murugesapillai et al. 2014). b In a magnetic tweezers setup, DNA tethered between a labeled paramagnetic bead and a functionalized cover slip is held at constant magnetic force and the extension is recorded using a CCD camera. Magnetic tweezers combined with fluorescently labeled proteins (green) allows visualization as well as quantification of protein binding. (Adapted from Skoko et al. 2004 and Xiao et al. 2010). c In a dual trap optical tweezers setup, DNA tethered between labeled polystyrene beads is extended and released. Fluorescently-labeled molecules (green) interact with the DNA and their binding can be visualized. (Adapted from Heller et al. 2014). d Atomic force microscopy is used to visualize protein–DNA complexes. The reflection of the laser beam off the cantilever to detector is then converted into an imaging signal. (Adapted from Murugesapillai et al. 2014)
