Fig. 14.
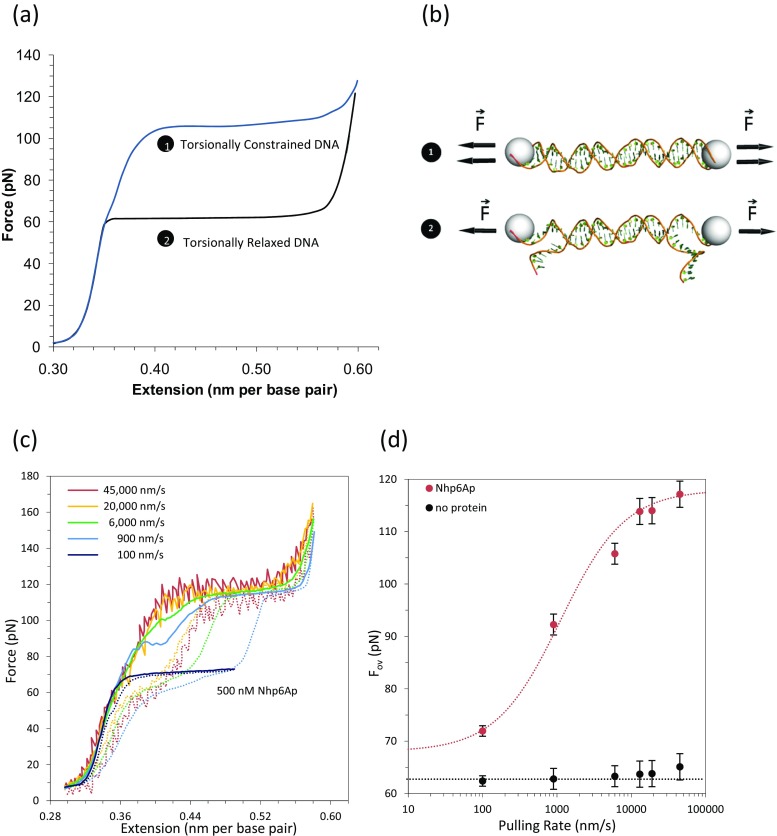
DNA torsionally constrained overstretching transition and the kinetic analysis of Nhp6A protein binding to DNA. a Torsionally constrained DNA is characterized by an overstretching force at 110 pN whereas torsionally relaxed DNA experiences the overstretching force at 65 pN. At high pulling rates, DNA in the presence of HMGB proteins displays an overstretching transition at forces comparable to those expected for torsionally constrained DNA. This result is interpreted as evidence that bound HMGB proteins block DNA unwinding. At low pulling rate the increase in overstretching transition due to protein binding occurs at ∼75 pN. b Schematic of torsionally constrained DNA (1 unable to rotate), and torsionally unconstrained DNA (2 able to rotate and unwind). c In the presence of HMGB proteins, DNA becomes torsionally constrained at pulling rates higher than the protein dissociation rate. d DNA overstretching force in the absence (black) or presence (red) of Nhp6A versus pulling rate, fitted to Eq. (8) to estimate the dissociation rate, k off. (Adapted from McCauley et al. 2013)