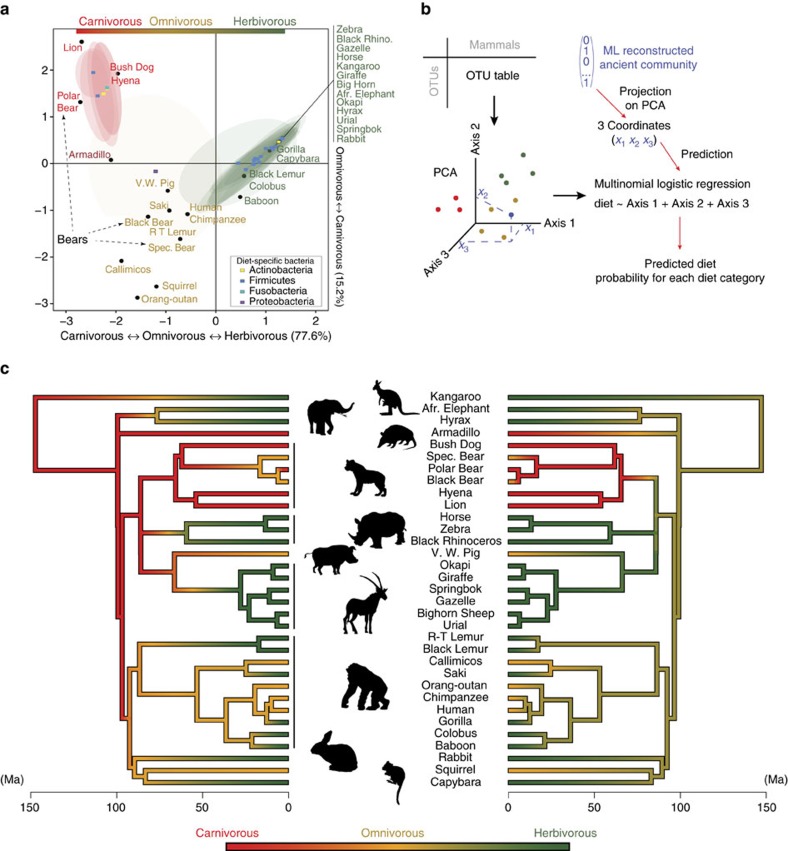
Figure 2. Gut microbiomes predict extant and ancestral mammalian diets.
(a) Diet-correlated bacterial lineages (squares coloured by phylum) and mammals (black dots) separated by dietary distances are jointly projected in the same ordination space using the OMI59 mapping procedure (see Methods). Predicted niche breadth59 of a bacterial lineage is depicted with an ellipse. The time period selected to define bacterial lineages is ∼300 Myr ago. Bacterial niche breadths show that many bacterial groups are herbivore or carnivore specific. However, no bacterial lineage has a niche breadth encompassing omnivores only (omnivores are in brown). (b) Workflow for microbiome-based prediction of ancient diet. (c) Inference of ancient mammalian diets. Left: trait-based reconstruction using 1,534 mammals34. Right: Microbiota-based reconstruction with the 33 mammals under study. At each ancestor, the probability vector for each diet category is transformed into a linear variable bound between 0 (carnivorous) and 1 (herbivorous). Between two ancestors, diet is assumed to evolve linearly. Animal images courtesy of Julien Renaud.