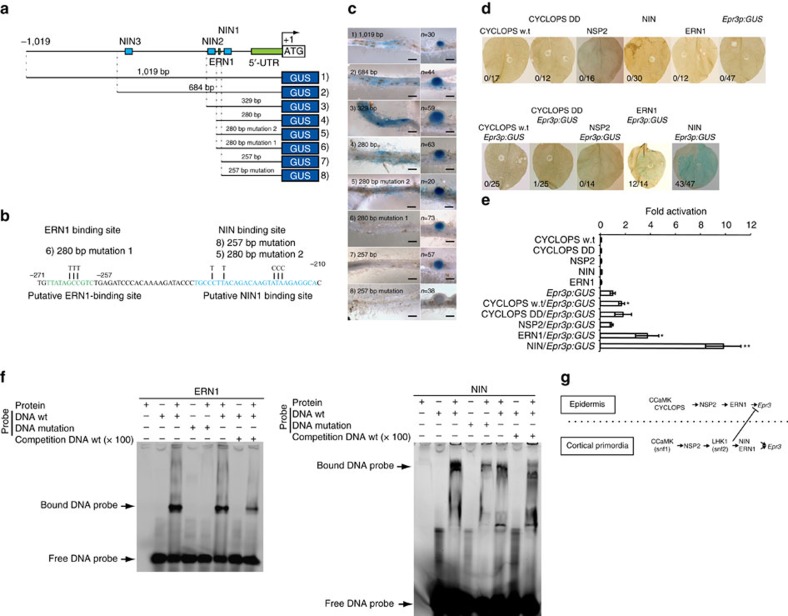
Figure 7. Epr3 promoter analysis in Gifu.
(a) Overview of the Epr3 promoter region and the analysed deletion series. Positions of three putative NIN-binding sites and one putative ERN1-binding site are indicated. (b) Nucleotide changes in the mutated putative binding sites for ERN1 and NIN. (c) Gifu plants transformed with pEpr3:GUS promoter deletions were inoculated with R7A and assayed at 4–10 dpi. Panels show Epr3 expression in epidermal and cortical cells. Scale bars, 0.5 mm. (d) The pEpr3:GUS, and/or constructs for overexpression of Cyclops, Cyclops DD, Nsp2, Ern1 and Nin were infiltrated into N. benthamiana leaves. Upper row shows single infiltration, and bottom row is co-infiltration of pEpr3:GUS together with CYCLOPS, CYCLOPS DD, NSP2 or NIN. After 3 days incubation, these leaves were stained by X-gluc, and destained. Numbers indicate fraction of GUS-positive leaves. (e) Levels of GUS activity induced by pEpr3:GUS in N. benthamiana leaves after 3 days infiltration. Panel shows fold change compared to pEpr3:GUS single infiltration. Error bars are s.e. * or ** (P<0.05 or P<0.01, Student's t-test, n=4–5) indicates significant difference compared to pEpr3:GUS single infilutaration. (f) EMSAs were performed using His-tagged ERN1 and truncated NIN (residues 520–878) together with putative ERN1- and NIN-binding DNA sequences. Sequences of DNA wt and mutant versions of the Epr3 promoter corresponds to b, and DNA probes were FAM-tagged. Competition DNA is 100 times concentration of wt DNA without FAM. (g) Model of Epr3 induction. In the epidermis, ERN1 is sufficient for Epr3 expression downstream of CCaMK, CYCLOPS and NSP2. In the cortical primordia, both NIN and ERN1 induce Epr3 expression directly downstream of CCaMK, NSP2 and the LHK1 cytokinin receptor. Cytokinin signaling through LHK1 has a negative effect on epidermal Epr3 expression possibly involving the mechanism regulating infection thread formation43.