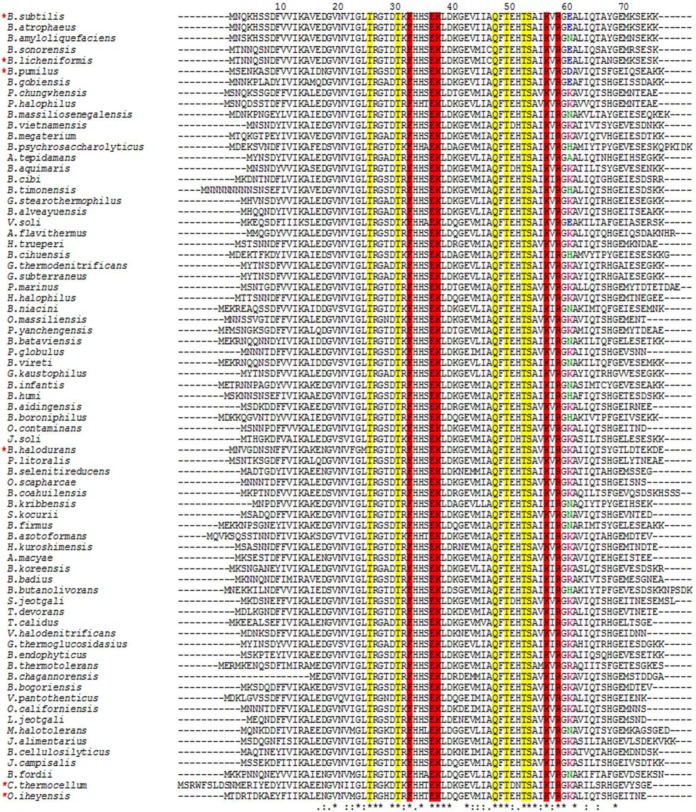
FIG 4.
Sequence alignment of TRAPs from different bacterial species. The amino acid sequence alignment was generated using Clustal Omega for 73 TRAP sequences from bacterial species (39). The residues involved in tryptophan binding (yellow) and RNA binding (red) are highlighted. Acidic residues at position 60 are shown in blue, basic residues are displayed in fuchsia, and uncharged residues are shown in green. Asterisks show positions that are 100% conserved, colons show positions with conservation of strongly similar amino acids, and dots indicate conservation between weakly similar amino acids. Numbers correspond to the B. subtilis TRAP sequence. Red asterisks at the left indicate the TRAPs examined for in vitro transcription attenuation with B. subtilis RNA polymerase (see Fig. 5).