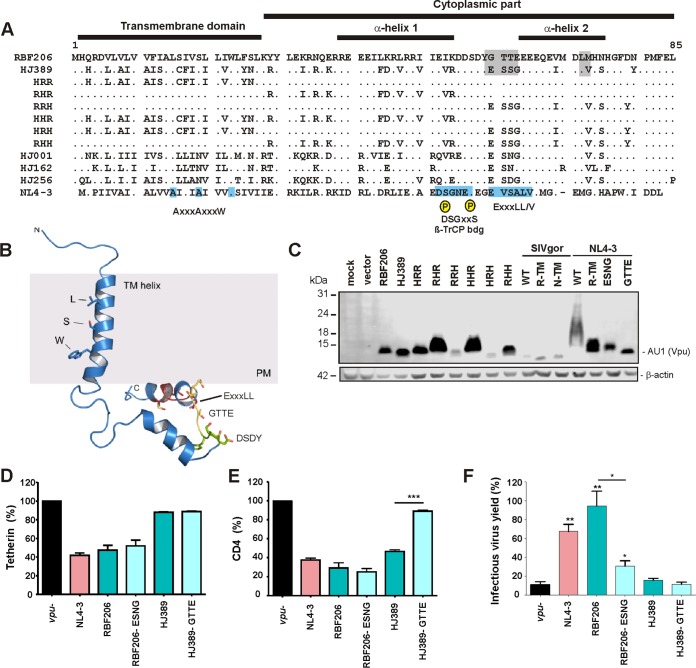
FIG 5.
Mapping of residues critical for the anti-tetherin activity of RBF206 Vpu. (A) Alignment of HIV-1 O Vpu amino acid sequences. The NL4-3 M-Vpu sequence is shown at the bottom for comparison. The TMD, the cytoplasmic part, α-helix 1, and important motifs and residues are indicated. The AxxxAxxxW motif residues in the TMD important for anti-tetherin activity of M-Vpus, the consensus DSGxxS β-TrCP interaction site, and the (E/D)xxxL(L/I/V/M) motif involved in targeting of tetherin for endosomal degradation and recruitment of AP-1 and AP-2 are indicated in the well characterized NL4-3 Vpu. (B) Localization of specific sequence motifs in the RBF206 structure. A structural model of RBF206 Vpu is based on homology modeling using the NMR structure 2N28 (85). The LxxxSxxxW motif within the transmembrane domain of RBF206, which degenerates from the typical AxxxAxxxW consensus, the cytosolic DSDY and GTTE motifs, and the ExxxLL internalization motif are indicated. (C) Expression of analyzed mutant RBF206 and HJ389 variants. HEK293T cells were cotransfected with constructs expressing the indicated Vpus or empty vector and analyzed by Western blotting. β-Actin was included as a loading control. (D and E). Effect of wild-type and GTTE/ESSG mutant RBF206 and HJ389 Vpu proteins on tetherin (D) and CD4 (E) cell surface expression. PHA-activated PBMCs were transduced with VSV-G-pseudotyped nef-defective HIV-1 NL4-3 IRES-eGFP constructs containing the indicated vpu alleles and analyzed by flow cytometry 2 days later. The levels of tetherin or CD4 surface expression in the presence of Vpu relative to that of cells transduced with the vpu-deficient control (100%) are shown. (F) HEK293T cells were cotransfected with an HIV-1 NL4-3 Δvpu Δnef construct, vectors expressing the indicated Vpu proteins, and a construct expressing human tetherin. Viral supernatants were obtained 2 days later and used to quantify infectious HIV-1 in the culture supernatants by infection of TZM-bl indicator cells. Values in all bar graphs represent averages (±SEM) from three experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.