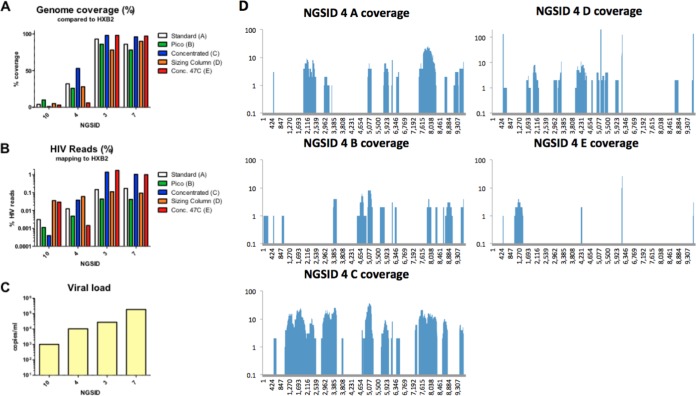
FIG 4.
Library preparation optimization. The trimmed NGS reads from HIV-SMART libraries were prepared by protocols A to E (A, the standard protocol; B, a protocol with the Pico SMART cDNA kit; C, a protocol with a nucleic acid concentrator; D, a protocol with a sizing column; E, a protocol that followed the nucleic concentration protocol [Conc.] in protocol C with reverse transcription at 47°C) and mapped to the HXB2 reference genome. (A, B) The genome coverage (A) and percentage of HIV reads (B) were calculated for this alignment by the use of CLC Bio software. (C) The viral load for each sample tested is plotted on a log scale. (D) The genome coverage plots for each position of the genome are shown for isolate NGSID 4, which showed a trend representative of the trends seen for all other isolates tested. y axis, number of reads; x axis, nucleotide position in the genome sequence.