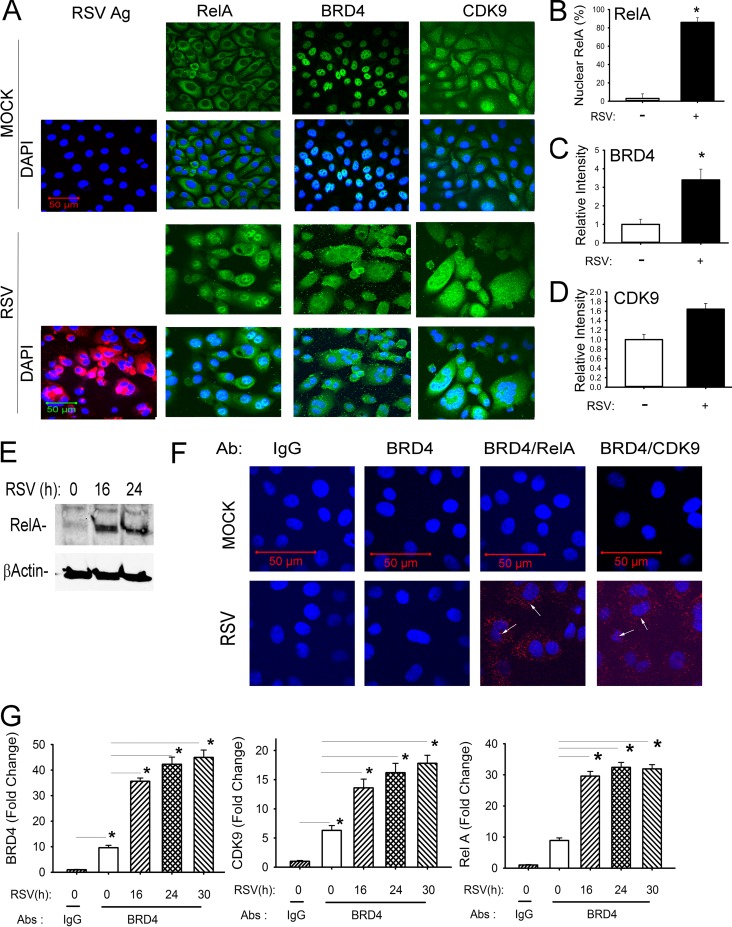
FIG 1.
RSV induces activation of the NF-κB/RelA-BRD4 complex. (A) Indirect immunofluorescence assays of RSV-infected hSAECs. hSAECs were exposed to sucrose (mock) or RSV (MOI of 1.0) for 24 h. After fixation, cells were stained with anti-RSV, -RelA, -BRD4, or -CDK9 Abs as indicated. Secondary detection was performed with Alexa 568 (red)-conjugated goat anti-rabbit IgG. Nuclei were counterstained with DAPI (blue). For each antigen (Ag), Alexa staining is shown in the top row; the merged DAPI image is shown in the bottom row. Images were acquired by confocal microscopy at ×63 magnification. (B) Quantification of RelA. Nuclear RelA was scored in 5 independent fields. Shown is percentage of RelA-positive cells (n = 100 for each). (C) Quantification of BRD4. Relative fluorescence intensity was quantified in 5 separate fields. (D) Quantification of CDK9. Quantification of CDK9 relative fluorescence intensity is presented as in panel C. (E) Activation of NF-κB/RelA in response to RSV infection. Nuclear extracts from uninfected (time zero) or RSV-infected (16 and 24 h p.i.) cells were prepared and analyzed by Western immunoblotting. (Top panel) Staining with anti-RelA Ab. (Bottom panel) β-Actin staining was used as an internal control. These data confirm nuclear translocation of RelA (A). (F) In situ proximal ligation assay of mock- or RSV-infected hSAECs (MOI of 1.0) for 24 h. Cells were fixed and subjected to PLA using IgG, anti-mouse BRD4 (anti-mBRD4) Ab alone, anti-mBRD4 plus anti-rabbit RelA, or anti-mBRD4 plus anti-CDK9 as indicated. Nuclei are counterstained in DAPI (blue); interacting proteins appear as red foci in both cytoplasm and nucleus (arrows). Images were acquired by confocal microscopy at ×100 magnification. (G) IP-SID-SRM-MS of BRD4 complex from control and RSV-infected hSAECs. Whole-cell lysates were immunoprecipitated using IgG or anti-BRD4 Abs. (Left panel) BRD4 abundance was detected by monitoring precursor-product transitions of 3 proteotypic peptides. Shown is the fold change in BRD4 signal abundance. (Middle panel) CDK9 abundance in BRD4 complexes was measured by SID-SRM-MS. (Right panel) RelA abundance. *, P < 0.05.