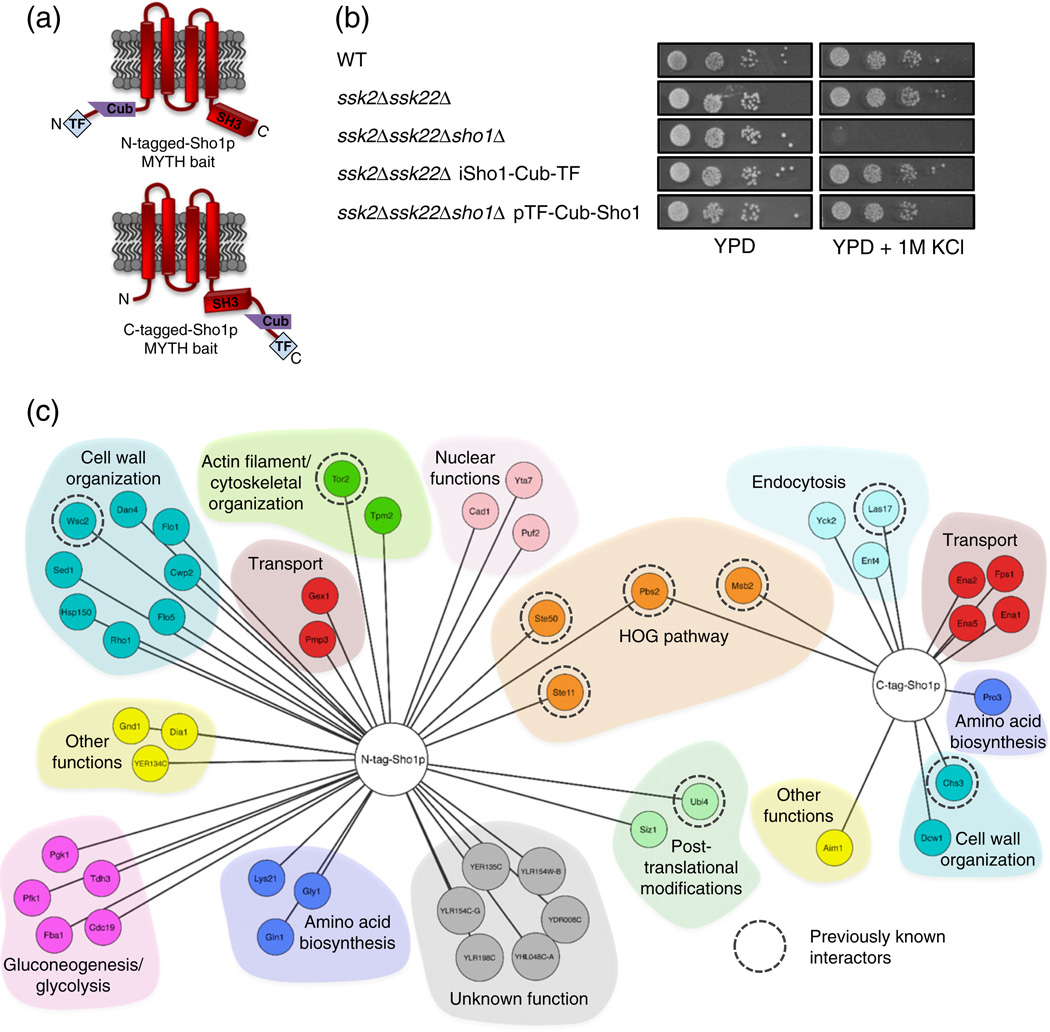
Fig. 1.
Application of Membrane Yeast Two-Hybrid (MYTH) to the Sho1p interactome. (a) Sho1p MYTH baits used in this study. The location of MYTH tag fusions (at the N- or the C-terminus of the protein) is indicated. (b) Sho1p baits are functional in the HOG pathway. Wildtype cells (WT) and deletion strains (as indicated) in the presence or absence of MYTH-tagged Sho1p bait were 10-fold serially diluted (from a starting OD0.05), spotted onto solid media, grown for 2 days at 30 °C, and imaged. All images are from the same plate. (c) Proteins identified as interacting protein partners of N-tagged (TF-Cub-Sho1p) and C-tagged (Sho1p-Cub-TF) Sho1p by the MYTH method. Proteins are grouped by colour according to function, and those found in previously reported physical or genetic interaction screens are indicated by a broken circle.