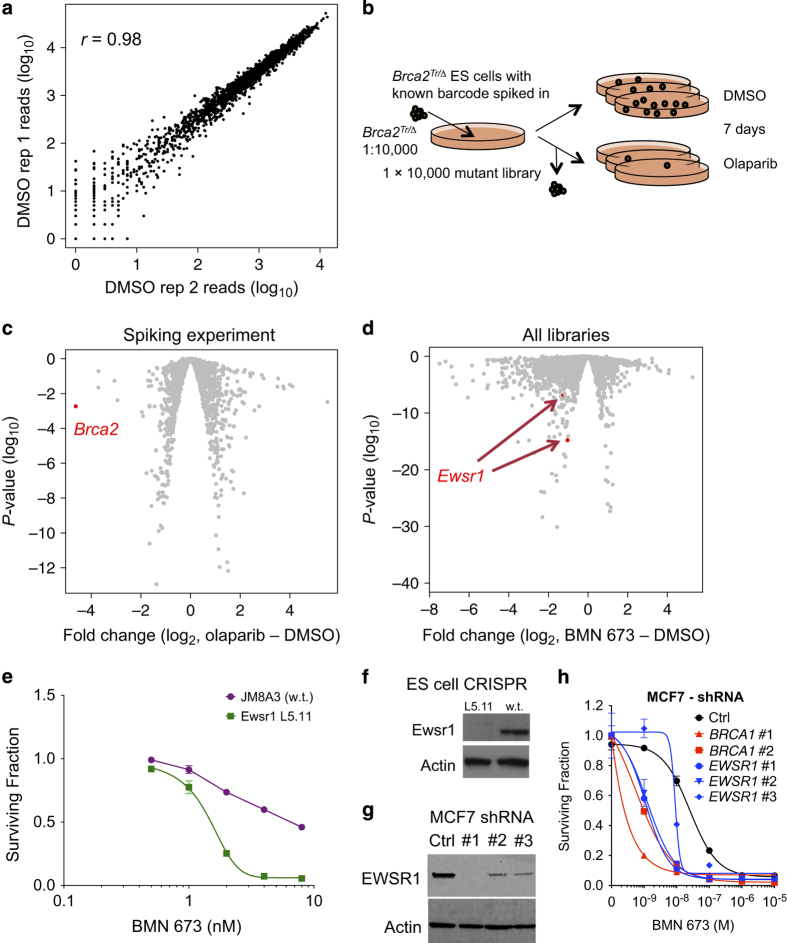
Figure 3. Technical validation.
(a) Concordance of read counts in two replicate DMSO-treated samples. (b) Schematic of spiking experiment. (c) Volcano plot of spiking experiment results. P values (exact test) and fold changes were calculated using edgeR. The point corresponding to the barcode in the spiked Brca2 mutant is labelled. (d) Volcano plot of barcodes corresponding to Ewsr1 integrations in all PB3 libraries exposed to the PARP inhibitor BMN 673. (e) Validation of Ewsr1 hit. CRISPR knockout ES cells (L5.11) are more sensitive to BMN 673 than parental JM8A3 cells. Surviving fraction is measured using CellTiterGlo after five days growth in the indicated drug concentration; mean and s.d. of five replicates is plotted. (f) Western blot demonstrating knockout of Ewsr1 in CRISPR mutant. (g,h) Knockdown of the human homologue, EWSR1, in MCF7 cells using shRNA causes BMN 673 sensitivity.