Abstract
For most of Medicine’s past, the best that physicians could do to cope with disease prevention and treatment was based on the expected response of an average patient. Currently, however, a more personalized/precise approach to cardiology and medicine in general is becoming possible, as the cost of sequencing a human genome has declined substantially. As a result, we are witnessing an era of precipitous advances in biomedicine and bourgeoning understanding of the genetic basis of cardiovascular and other diseases, reminiscent of the resurgence of innovations in physico-mathematical sciences and biology-anatomy-cardiology in the Renaissance, a parallel time of radical change and reformation of medical knowledge, education and practice. Now on the horizon is an individualized, diverse patient-centered, approach to medical practice that encompasses the development of new, gene-based diagnostics and preventive medicine tactics, and offers the broadest range of personalized therapies based on pharmacogenetics. Over time, translation of genomic and high-tech approaches unquestionably will transform clinical practice in cardiology and medicine as a whole, with the adoption of new personalized medicine approaches and procedures. Clearly, future prospects far outweigh present accomplishments, which are best viewed as a promising start. It is now essential for pluridisciplinary health care providers to examine the drivers and barriers to the clinical adoption of this emerging revolutionary paradigm, in order to expedite the realization of its potential. So, we are not there yet, but we are definitely on our way.
Keywords: Personalized/precise medicine, DNA microarrays, Next-generation genome sequencing (NGS), Genome-wide association studies (GWAS), CRISPR-Cas9 genetic-phenotypic screen, Molecular genomic decoding of phenotypic diversity, Omics-based tests, Molecular genetic biomarkers, Pharmacogenomics and pharmacogenetics
It is more important to know what sort of person [individuality] has a disease, than to know what sort of disease a person has —Hippocrates (460-370 BCE), and Sir William Osler (1849–1919)
Diathesis (Gk., signifying a constitutional predisposition toward a particular state or condition and especially one that is abnormal or diseased) … is nothing else but a chemical individuality … the factors which confer to us predispositions to and immunities from the various mishaps which are spoken of as diseases, are inherent in our very chemical structures — Sir Archibald Garrod (1857–1936)
1. Hippocratic origins of personalized medicine
Every patient is unique or distinctive, and the evolving field of personalized medicine (PMed) aims to ensure the delivery of the proper treatment to the right patient at the right time. It was Hippocrates (born c. 460 BC, island of Cos, Greece; died c. 375, Larissa, Greece), the Father of modern Medicine [1] (see Fig. 1), who first underscored the patient as the most important determinant of therapeutic efficacy. He had somehow recognized the central principle of PMed: human beings are innately (genetically) different from one another, and this individuality affects both their predisposition/susceptibility to disease and their response to therapeutics [2]. Accordingly, optimal clinical practice must entail a personalized or individualized approach to diagnosis and treatment. The ramifications of such a personalized approach and resultant ideas also extend into academic realms traditionally reserved for the social sciences, philosophy, ethics and religion, as well as jurisprudence, the discipline of what is just and unjust.
Fig. 1.
Marble statue of Hippocrates, ancient Greek physician considered as the Father of Western Medicine, in the city of Larissa in Thessaly, on the mainland of Greece. He envisioned the need for a Personalized Medicine.
Hippocrates advanced a rational, targeted therapeutic strategy. Treatment should be etiological (cause-oriented) rather than phenomenological (symptom-oriented) [Places in Man; v. VIII, 1995]. Therefore, it is necessary to inquire into and treat the etiology or cause [Nature of Man; v. IV, 1931] of why manifest symptoms come about [Ancient medicine; v. I, 1923] [3]. It is essential to first understand disease pathophysiology, which in turn requires the knowledge of human biology [Nature of Man; v. IV, 1931] [3].
Hippocrates was the first to highlight that there are critical natural nonuniformities and a great difference in the constitution of individuals: each person exhibits one’s idiosyncrasia (idiosyncrasy) [On Joints; v. III, 1928] [3]. Idiosyncrasy predisposes to or protects from particular diseases [The Sacred Disease (Epilepsy); v. II, 1923] [3]. The individuality that Hippocrates first identified is today understood at long last as an individuality at the level of the genome/epigenome, which encode the information that governs the human body’s composition, traits, disease predisposition, response to treatment, and prognosis.
2. Genomic structures weave a thread through modern medicine: gene therapy
Developments and breakthroughs in the basic sciences and biotechnology have in our day revolutionized medical research, including cardiovascular investigations, bringing about diversified advances many of which could not have been forecasted even 1–2 decades ago. Progress in computational methods, biomedical engineering, multimodal and multiscale imaging, molecular and cellular biology and genetic engineering, genomic sciences, proteomic technologies, and epigenetics are among the many advances that have speeded up the pace of biomedical research. Today we are aware that biology creates its structures—proteins, hormones, cells, tissues, organs and the like—by putting genes into intricately regulated action.
In our own species there are roughly 21,000 protein encoding genes (estimates continue to fluctuate), and genome-wide variation from one person to another can be up to 0.5% (99.5% agreement). Interestingly, cats have 90% of homologous genes with humans [4] and 50% of Drosophila melanogaster (“fruit fly”) protein sequences have mammalian homologs [5], while about 75% of known human disease genes have a recognizable match in the genome of D. melanogaster [6]. Evidently, all organisms have a common ancestor and therefore share many components of their genomes. Individual genes function collectively as the parts of a programing language for the formation of an enormous variety of products of diverse form and function. Organisms cast themselves in many distinctive shapes and amazingly disparate species by using much the same set of genes “programed”—in conjunction with “environmental/epigenetic factors—to become activated in precise progressions and operational integrative patterns, in health and disease.
2.1. The restructuring of genetics and its convergence with medicine
Genetics nowadays has established itself not only just as a specialty of medicine but also as a thread throughout all of health care [7] and is now embarking on the most ambitious and inventive phase of its existence. By 1965, when I was being taught different aspects of molecular biology—a melding of biochemistry and genetics—by the Medicine Nobel Laureates Severo Ochoa and Baruch Benacerraf at NYU School of Medicine, molecular biology had laid bare the basic secrets of genetics residing on the DNA double helix consisting of antiparallel helical strands, with complementary bases. Without the ability to manipulate genes, however, the understanding was more theoretical than operational.
In the 1970s, this situation was transformed by the recombinant DNA technology, which led to transgenic organisms and gene therapies. A variety of enzymes were discovered, made by bacteria, which allowed DNA manipulation as needed. Bacteria make restriction enzymes, which cleave DNA at specific sequences and function as defense against invading viruses, and ligases, which join DNA fragments. With these and other tools—which today are available commercially—it became possible to cut and paste DNA fragments from one genome to another as desired and using a variety of recombinant DNA technologies introduce them, through a vector, into cells to supplant variant defective human gene(s) directly and thereby enable endogenous production of desirable compound(s). Such nanoscale therapeutic interventions can result in as impressive outcomes as conventional interventional cardiology procedures [8].
Generally, gene transfer entails adding a functional gene (a transgene) into a recipient’s body, correcting a dysfunctional gene to wheedle tissue production of desirable compounds endogenously, or altering the expression of a naturally occurring gene; accordingly, gene transfer and gene therapy are not synonymous. Today, DNA fragment cloning procedures altering gene expression also allow investigators to reproduce unlimited quantities of specific DNA molecules and, along with the polymerase chain reaction (PCR) that allows one to directly amplify a specific DNA sequence and the invention of DNA sequencing—the process of determining the order of bases in a segment of DNA—have led to mapping and detailed understanding of many individual genes and coordinated, integrative gene networks.
In consequence, in our time, it is acknowledged that virtually all human ailments/disorders excluding random trauma, have some genetic component. Since certain portions of genetic information are unique to each individual, genetic variation/dissimilarity has some bearing on every aspect of human biology, development, morphomechanics, and miscellaneous adaptations. Consequently, identifying and understanding human genetic variation should play a pivotal role in promoting health and fighting against disease. This paradigm-changing realization has generated exceptional opportunities for advancing medical science and holds promise for upgrading clinical care and health outcomes. It has fostered the advent of a personalized precision medicine [9] nearly two-and-a half millennia after it was envisioned by Hippocrates [2], notwithstanding the technological constraints of his times.
3. Human diversity brings about the personalized medicine/cardiology paradigm
The advent of PMed came about as the inevitable outcome of momentous scientific and technological innovations. To begin with, great strides toward patient-specific pathophysiological and diagnostic investigations, including the outcomes of therapeutic interventions, supplemented spectacular advances in computer-assisted numerical simulations of normal and pathological cardiac fluid and myocardial mechanics, and hemodynamics [10–24]. Such simulations employ data-rich modeling of dynamic 3D “functional imaging” geometry from multimodal in vivo imaging and solid-state multisensory catheterization studies, in experimental animals and in patients being evaluated for cardiac disease and effectiveness of therapeutic procedures [10,25].
3.1. Patient-specific cardiovascular dynamics simulations using multimodal measurements
In digital computer simulations multimodal cardiac images and catheterization measurements become numerical data, so they can be subjected to mathematical manipulations that correspond to the physics of the cardiomechanics processes (e.g., right and left ventricular systolic contraction and ejection [19,22,26–28], diastolic relaxation and filling [29–34]) that are being simulated. Digital computer simulation works well as long as we can accurately reduce the process/phenomenon under investigation to a numerical description for mathematical manipulation. It then becomes numerical data that can be acted upon by powerful algorithms comprising a massive set of arithmetic operations or sequences of arithmetic and logical steps; these operations can be executed swiftly on powerful computers. Anatomically detailed models of the ventricles, including myocardial fiber orientations and laminar structure, have been used to reconstruct the dynamic, patient-specific, morphomechanical behavior of the whole organ working in situ, under diverse conditions of health and disease [10–24].
3.2. DNA variants and personalized strategies for diagnosis/treatment of genetic diseases
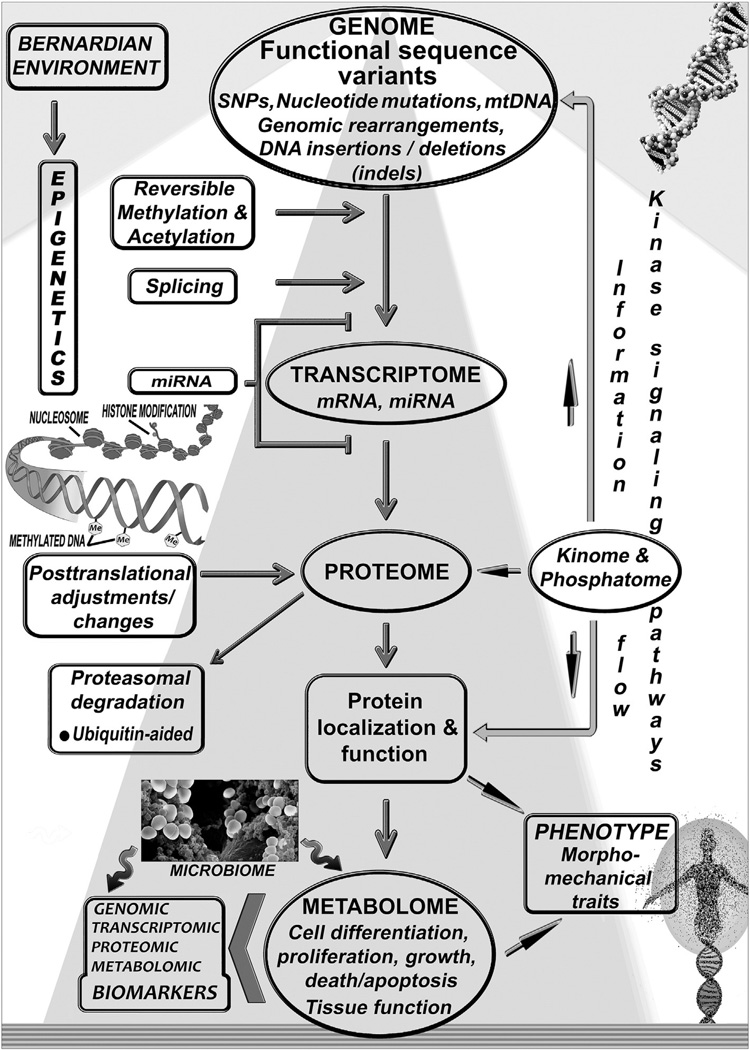
State-of-the-art developments in PMed issued from the Human Genome Project (HGP) in 2001 and the International HapMap Project in 2005 [35]. The HapMap is a catalog of common genetic variation, or haplotypes, in the human genome; a haplotype is a collection of alleles arranged linearly along a person’s DNA molecule. In 2010, the third phase of the HapMap project was published; HapMap data have accelerated the search for genes implicated in common human diseases [36]. These developments hinge on technology and tools created through the HGP; they delineate an individual patient’s essential biology—DNA, RNA, or protein (see Fig. 2)—which, taking everything into consideration, predisposes to or results in disease. They also subserve efforts to characterize the genomes of animal models used extensively in genomic research, including mice, roundworms, and fruit flies.
Fig. 2.
The genome integrates intrinsic and environmental signals. The Bernardian environment and epigenetic regulatory factors influence through (bidirectional) information flow and signaling pathways variable human gene transcription and expression, while protein levels and function are affected by various posttranslational adjustments. Epigenetic genome modifications allow the stable propagation of gene activity states from one cell generation to the next; since epigenetic states are reversible, they can be modified by environmental factors, which may contribute to modifiable changes between normal and abnormal phenotypes. Interfering with the activity of factors that modify the chromatin state can possibly affect the expression of unwanted genes. The human organism is effectively a “supraorganism,” an assemblage of human and microbial cells and genes and thus a blend of human and microbiome traits, including the metabolome. The relationship between different “-ome/-omics” components, protein levels, post-translational modification, protein localization and activity, leading to effects in cells and tissues and affecting phenotypic traits, is shown schematically in this general overview.
Many cardiovascular genetic disorders occur in high frequency in the population, ostensibly because their life-threatening injurious effects typically are delayed well beyond reproductive-age onset. The sequencing of the human genome has allowed linkage of a growing number of diseases—including cardiac arrhythmias [37–39]; ischemic heart disease [40–43]; assorted cardiomyopathies (see Table 1; the data listed are a tip of the iceberg) [35,44–46]; congestive heart failure [47,48]; and hypertension [49–51]—to numerous genetic variants (Fig. 2) and multicomponent/multifactorial genomic abnormalities [10,35]. An exemplary demonstration is the 1985 Nobel Prize-receiving discovery by Michael Brown and Joseph Goldstein [52,53] that mutations of the low density lipoprotein (LDL) receptor gene family affecting the LDL cell-surface receptor—an 839 amino-acid protein that mediates the endocytosis of cholesterol-rich LDL—cause hypercholesterolemia, development of atherosclerosis, and early-onset myocardial infarction. This finding has resulted in LDL-cholesterol lowering therapies that lessen cardiovascular-event risk.
Table 1.
Genes commonly implicated in assorted cardiomyopathies in descending order of frequency
| Gene | Protein name | |
|---|---|---|
| % of HCM associated with mutation of this gene | ||
| 40% | MYH7 | Myosin-7 |
| 40% | MYBPC3 | Myosin-binding protein C, cardiac type |
| 5% | TNNT2 | Troponin T, cardiac muscle |
| 5% | TNNI3 | Troponin I, cardiac muscle |
| 2% | TPM1 | Tropomyosin alpha-1 chain |
| % of DCM associated with mutation of this gene | ||
| 20% | TN | Titin |
| 6% | LMNA | Lamin-A/C |
| 4.2% | MYH7 | Myosin-7 |
| 3%–4% | MYH6 | Myosin-6 |
| 2%–4% | MYBPC3 | Myosin-binding protein C, cardiac-type |
Table prepared using data from pertinent chapters of GeneReviews [Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2015].
Reproduced from Pasipoularides [35] with permission of the Journal of Cardiovascular Translational Research.
However, most currently available drugs target a single gene or protein disregarding the dynamics of highly complex biomolecular networks that collectively drive gene expression and cellular/organ functions. It then follows that most do not treat common multifactorial disorders, while diagnostics and biomarkers available in tandem are unable to predict tissue-specific cellular reactions to genetic and epigenetic alterations—we are not simply the sum of our genes—as well as drug effects in individual patients and disparate patient groups. Clearly, uncovering the precise risk factors for multifactorial disorders will require a staggering amount of data from a huge number of individuals. Accordingly, substantial potential benefits should accrue by applying personal whole genome analysis of individual patients for personalized treatments. In addition, the discovery/development of next-generation drugs should be shifted from the conventional “one-size-fits-all” approach to gene network-associated disease mechanisms integrating genome science and network/systems-biology dynamics.
In this context, significant advances have been made in mapping many molecular pathways by which genetic variants or mutations manifest themselves as cardiovascular disease [54]. These advances have enabled development of diagnostic tools that can set apart well-defined subtypes of what had previously been considered as a single disease, together with molecular agents that target each subtype, thus providing the basis for effective personalized cardiology (PCard) (Figs. 3 and 4). The utility of whole-genome sequences has stimulated the development and application of genomic technologies, such as microarrays and next-generation genome sequencing (NGS), which are reviewed in Appendix A.
Fig. 3.
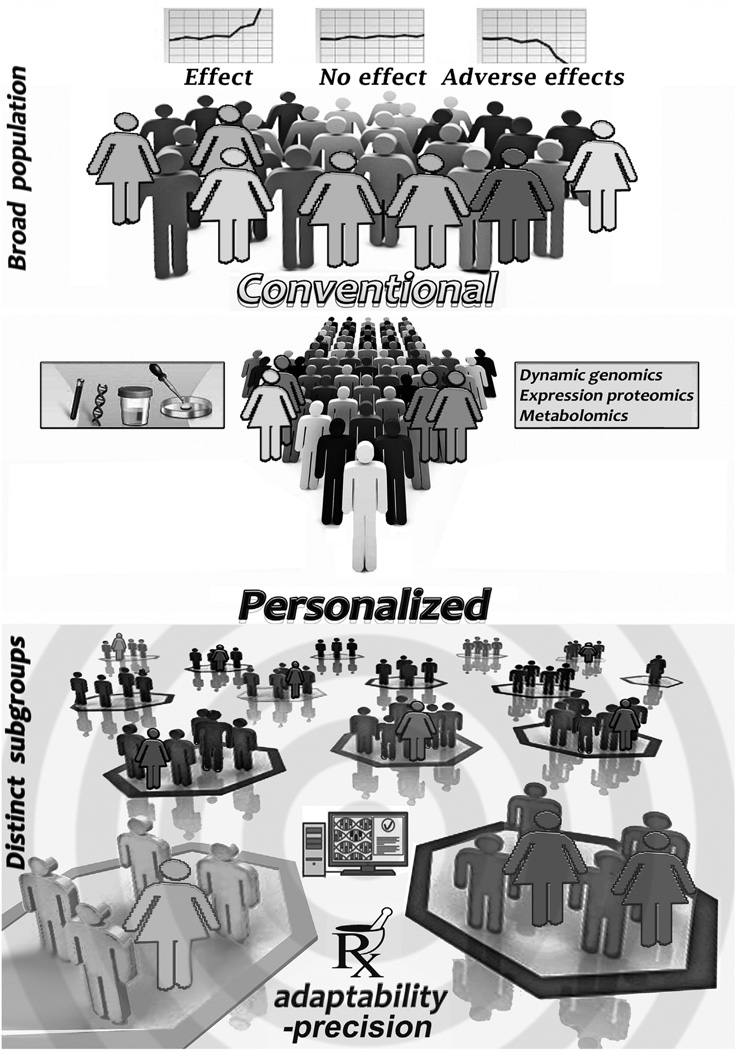
Substantial interindividual variability can emerge in the clinical response to accepted treatments for acute and chronic diseases. Genetic, metabolic and med history data can split patients into smaller groups, allowing treatments to be tailored with precision to the specific subgroups, so as to augment efficacy and minimize undesirable adverse effects.
Fig. 4.
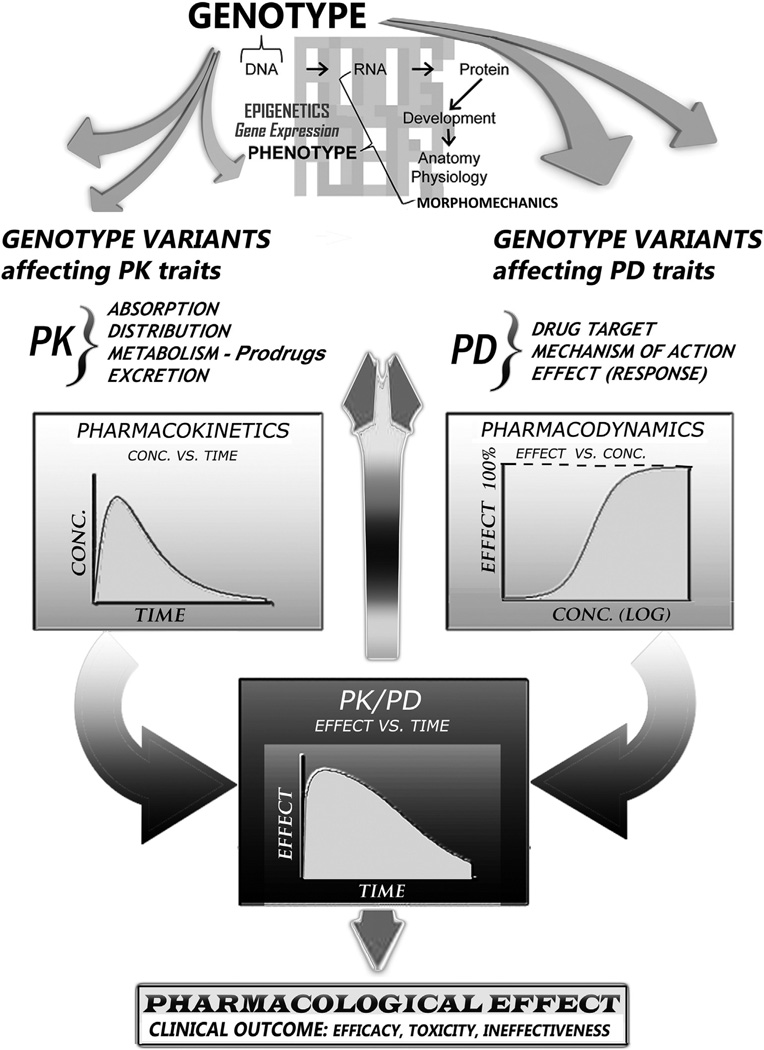
Pharmacokinetics (PK) studies the time course of drug absorption, distribution, metabolism, and excretion and then applies resulting principles to the safe and effective therapeutic management of drugs in an individual patient Pharmacodynamics (PD) concerns the relationship between drug concentration at the site of action and the resulting response, including the time-course of therapeutic and adverse effects. The effect of a drug present at the site of action is determined by its binding with a receptor; receptors present on cardiac muscle affect, e.g., the intensity of contraction. Genotypic and epigenetic variability affect drug response in several ways involving: varying receptor density on the cell surface; mechanisms by which a signal is transmitted within the cell by second messengers; or, complex factors controlling gene expression and protein production. This multilevel PD regulation results in variation of sensitivity to drug effect(s) from one individual to another and, combined with PK governing the amount of time that the drug is present at its action sites, also determines enhancement of or tolerance to drug actions and clinical outcome.
3.3. Gene function prediction directly from genomic sequence data
In parallel with the mapping types of analysis, the availability of enormous quantities of genomic sequence data, accruing from the human and other genome sequencing projects, has motivated computational investigations of gene function prediction directly from genomic sequence data, bringing into play statistical analysis and automated pattern recognition software—e.g., the online Genome Sequence Annotation Server (GenSAS), https://gensas.bioinfo.wsu.edu/. The archetypical rule in predicting gene function by means of a computational approach is guilt/honor by association, asserting that qualities of one item are inherently qualities of another simply by way of an inapt association: if two genes are structurally related, then they may express comparable functions.
However, such designations/attributions are fraught with problems. Specifically, a “gene” is not an appropriate unit for the purpose of predicting function, given that the vast majority of human genes are alternatively spliced and most are spliced in a tissue-dependent manner resulting in disparate expression patterns/profiles. We make protean use of our genes; in different tissues a single gene may generate multiple transcript variants and protein isoforms with functions as different as those of different genes, thereby greatly expanding the repertoire of the 20,000 or so genes in the human genome. Additionally, chemical epigenetic modifications of DNA that do not affect the primary base-pair sequence also affect gene expression. Moreover, close scrutiny of the term “function” in the context of genomic biology shows that genes act in complex interdependent ways to bring about cellular processes, and do not function in isolation.
Conceivably, however, computational gene function prediction, recognizing the parts of the genomic DNA sequence that contain pertinent biological features, might expedite and assist in the many-sided process of inferring regulatory multicomponent networks of genes interacting in various cellular processes. And a host of increasingly sophisticated algorithms can predict whether a mutation is likely to change the function of a protein, or alter its expression [55].
4. Diagnostic DNA sequencing systems
The translation of vast genomic data/information into metabolic function, normal and abnormal physiology, and disease potential, is a great challenge of PMed. Achieving this goal means confronting several challenges in correlating genome sequence information and gene function with normal/disease phenotypes, identifying molecular metabolic engines that drive cellular processes, delineating genetic regulatory networks, and understanding normal and diseased tissue and organ integrative function beyond individual cells. Techniques involved in these investigations include (but are not limited to) immunohistochemistry, in-situ hybridization, mutational analysis, protein analysis, cell culture and isolation, PCRs, expression profiling, blotting and microarrays.
Sequencing DNA and RNA molecules in human normal and disease samples is an integral component of a wide variety of PMed applications. As DNA sequencing technology progresses, it affords researchers and clinicians a variety of tools to probe the order of nucleic acid residues in genomes in great depth, a precondition for subsequently investigating how genome sequence variants and their expression patterns underlie phenotype and disease. Powerful innovative methods of personal genome studies are opening up new insights into genomic complexity and PMed, and permit high-throughput sequencing to be not only a discovery activity but also a practical assay for hypothesis testing. A few selected high-throughput sequencing methods are considered in Appendix A.
5. From traditional trial-and-error to personalized precision medicine
Because of poor cost-effectiveness of the—often only modestly efficacious in heterogeneous patient groups—population-biased “one-size-fits-all” approach, attention is again turning from standard-treatment and conventional drug development, to Hippocrates’ ancient paradigm, highlighting personalized (patient-specific) chemical molecular considerations à la Garrod—see Epigraphs. Currently, genetic/genomic and related developments allow precise diagnosis, not based on phenomenology alone, and precise treatment of an expanding number of cardiovascular diseases much earlier and with greater accuracy than ever before [10, 35,54,56,57]. Therapy can now be customized, optimizing the efficacy of drug treatments while minimizing side-effects (see Fig. 3).
However, progress in realizing the clinical promise of PMed has been slower than hoped. Paradigm change rarely happens quickly. Today, for the most part, practicing cardiologists continue—as do most physicians [58]—to carry out conventional medicine. Therefore, the inherent variability among different individuals has a huge adverse effect on the quality and cost—collectively, billions of unintentionally wasted dollars—of national healthcare. It is exceedingly rare for a drug to be efficacious for everyone at all times (Fig. 3).
Successful pharmaceuticals deliver the looked-for response in the range of 50–75% of cases, and the percentage of patients in whom a diagnosis has been made on the basis of customary clinical evaluation but for whom standard drug treatment is ineffective ranges from 38% to 75% [59]. For some cardiovascular drugs, such as statins routinely used to lower cholesterol, as few as 1 in 50 may actually benefit [60,61].
Such statistics confirm the pressing need for suitable diagnostic tests to anticipate who should and who will likely not respond to prospective treatments, considering that many drugs (including notably, in a cardiological context, the statins) have “pleiotropic” (from the Gk. words pleion, signifying more, and tropos, connoting direction or way) effects, which may be beneficial, neutral, or even undesirable and acknowledged as adverse side-effects. For instance, cholesterol-independent beneficial pleiotropic effects of statins have to do with improving or restoring endothelial function, attenuating vascular and myocardial adverse remodeling, decreasing oxidative stress and vascular inflammation, enhancing the stability of atherosclerotic plaques, and inhibiting the thrombogenic response [62,63]. Many of these pleiotropic effects are mediated by inhibition of isoprenoids, which serve as lipid attachments for intracellular signaling molecules [35,36,129].
Imagine receiving a full diagnosis from a simple peripheral blood sample, or the capability to tailor the precisely corrective treatment to an individual patient’s genetic makeup, taking into account specific susceptibilities to side-effects. Today’s informatics technologies allow sophisticated websites to precisely and speedily tailor their offerings to a prospective buyer’s specific profile and history. It remains to be seen whether genetic testing data-processing could reach this level of personalization in the not-too-distant future, or whether providers may prove too reluctant to move away from the “one-size-fits-all” paradigm revolving around “standards of care,” despite genomic multi-disciplinary scientific advances that are forging ahead.
What if we could apply a patient’s genetic information to gain insights into the genetic variations of diseases and expedite drug development, to create more precise therapies? This is the essential goal of personalized precision medicine. Auspiciously, regulatory agencies such as the FDA have started to recognize the importance of individual patient responses; cf. FDA’s “boxed warning” on clopidogrel—clopidogrel (Plavix) is a prodrug that requires biotransformation by cytochrome P450 (see discussions in Sections 9 & 9.1) to generate its active metabolite [64]—FDA’s warning addresses the need for pharmacogenomic testing to identify patients’ atypical clopidogrel metabolism and ensuing risk for a suboptimal antiplatelet response [65,66].
6. Molecular genomic decoding of phenotypic diversity
Deciphering the functional/morphomechanical impacts of molecular variants in the human genome entails ascertaining their effects on gene expression. At present, molecular biologists are at work figuring out the function of all genes and associating DNA attributes with individual phenotype as well as medically relevant human characteristics. In contrast to the conventional trial-and-error medicine, personalized precision medicine exploits sophisticated diagnostic testing to identify accurately the precise disorder/abnormality. To come to the most suitable treatment and determine right prescription dosages, the PMed approach takes into account the individual patients’ unique genotypic biochemistry and resulting physiologic capacity to metabolize particular medications (see Figs. 3 and 4).
Implicit in the PMed approach to clinical practice is Aristotle’s portrayal of (patho) physiology/morphomechanics as the exquisite matching of form and function, down to the molecular level. For Aristotle (born 384 BC, Stagira, Greece; died 322 BC, Chalcis, Greece), living organisms were exquisite assemblies of systems; of organized structures and mechanisms. Aristotle’s idea was that heredity was the flow of information, a river of code embodying messages to create materials and formulate functions, which moved from fertilized egg to the embryo—it was the system that carried across generations the instructions to mold an embryo.
6.1. Variable gene expressivity and regulatory controls
Phenotypic diversity among individuals is a result of variations in DNA sequences as well as of environmental effects, including external triggers or random chance effects on genes that may have variable “expressivity” or incomplete (reduced) “penetrance” [67]. Reduced penetrance and variable expressivity are probably caused by a combination of genetic, environmental, random chance, and lifestyle factors; they imply that the probability that an individual inheriting disease-genes will actually manifest the disease phenotype may be <1.
For instance, the exhibited features of Marfan syndrome vary extensively [35,68]: some individuals have only mild symptoms (such as being tall and thin, with long fingers), while others experience life-threatening complications as well, involving the heart and the aorta. Although the features are highly variable, most persons with this disorder have a mutation in the same gene (fibrillin-1, FBN1), which exhibits variable expressivity. Fibrillin, the product of FBN1, is a glycoprotein constituent of microfibrils that provides a scaffold for the deposition of elastin; it is especially abundant in the connective tissues of the aorta. Because of incomplete penetrance, one cannot conclude that an unaffected person has inherited a normal copy of the gene. Correspondingly, reduced penetrance can render it challenging to interpret a patient’s family medical history or predict the risk of passing a genetic condition to future generations.
Genes can be regulated, i.e., turned “on” and “off” at proper times and in appropriate contexts by particular cues/signals coming from outside the genome and interacting with controlling promoter, operator, and silencer DNA segments/“switches” located alongside the genes; this allows genes to adapt their function to their changing “environment,” and to specify the unique and modifiable morphomechanical features of distinct cells, tissues and organs, each specialized to perform different bodily functions [69]. These bodily components, which all developed from the original totipotent zygote, cannot have any formative change in their phenotype—a novel phenotypic state—without an altered genetic integrative function pathway. Details of how their genes/DNA might specify during embryogenesis and subsequent stages of life the development of complex structures found in humans and other higher organisms in health and disease remains still a major opaque/unsolved problem. Regulatory mechanisms controlling the expression of many of the traits involved in cardiological disorders/disease must commonly be polygenic in nature, because they implicate complex aspects of the phenotype, including complexly responsive tissues and organs of the integrated cardiovascular system.
The amount of transcript of each gene can be treated as a phenotypic trait, since it reflects changes in protein function more reliably than DNA markers. Gene expression profiling, including use of serial analysis of gene expression (SAGE) [35,70,127], represents a potent tool for exploring functional genetic variation using RNA molecular genetic markers. SAGE generates complete expression profiles of tissues of interest; it involves the construction of total mRNA libraries which enable a quantitative analysis of the whole transcripts expressed or inactivated at particular steps of cellular/tissue activation.
6.2. Characterization of global sets of biological molecules: Omics-based tests
Although analysis of single types of biomolecules has proven extremely useful in understanding many biological phenomena, the parallel large-scale investigation of DNA, RNA and proteins exposes new perspectives in the interpretation and modeling of the complexity of normal and disease processes. As a result, fresh scientific disciplines with the suffix “-omics” have appeared, encompassing multiple molecular disciplines that entail characterization of biological molecules such as DNAs, RNAs, proteins, and metabolites, and developing a new understanding of the molecular and genetic basis of disease [71]. In these nascent fields, recent advances in the preparation, identification and sequencing of DNA, RNA, and proteins, and in large-scale digital molecular and phenotypic data storage and analysis, are bringing about a revolution in our scientific understanding of phenotypic diversity among individuals.
Omics-based tests yield complex high-dimensional data; such data are generated through measurement of many more predictor variables per sample than the total number of individual samples used to generate the dataset; datasets typically result from measurements of hundreds-to-thousands of molecules in a relatively small number of individual samples. By measuring, in each patient, thousands of genetic variations, mutations, or changes in gene and protein expression and activity, clinical investigators are identifying previously unknown, molecularly defined disease states and searching for complex biomarkers that predict responses to therapy and disease outcome. The analysis of such high-dimensional datasets requires appropriate computing power and statistical methods [72]. The accruing new understanding is starting to shape the ways in which diseases are managed and how new medicines/treatments and accompanying tests are being developed and exploited.
Auspiciously, over the past decade, NIH budgets have aimed toward investigations into microRNAs, gene-environment interactions, epigenetics, and translational medicine. In a turn from mapping to function, the post-HGP NIH has sponsored new research. Across numerous Reports and Strategic Plans, it has listed the resolution of the roles that genes play in diseases that exhibit disparities/incongruence among individual patients as a high priority of trans-Institute funding (http://www.fda.gov/downloads/ScienceResearch/SpecialTopics/WomensHealthResearch/UCM334959.pdf; http://www.aascu.org/WorkArea/Download Asset.aspx?id=17548).
7. Molecular pathology and diagnostics of disease
Without biological perceptiveness we cannot hope to cure individual diseases/disorders. It is constructive to consider that each human individual consists of 100s of trillions of cells, a large percentage of which are symbiotic bacteria (microbiome, encompassing many scores of bacterial species with their myriad beneficial molecular metabolic products; see Fig. 2) located primarily in our gut (http://dx.doi.org/10.1101/036103). We are composed of approximately 210 distinct cell types, which contain 20,000–25,000 genes, 10 million proteins (including antibodies) and 2,000–3,000 metabolites. Each virtual/representative nucleated human cell contains many billion RNA molecules and 6 feet—if stretched-out completely—of DNA containing 6,400,000,000 base pairs. Nucleic acids encode cell structure-function information.
Most genes encode proteins; when a gene is being expressed, its product is being made. The basic science discipline of Molecular Pathology is vital for PMed. It uses molecular techniques for the characterization of genetic lesions (DNA) in cell-nuclei, gene expression (protein, RNA), gene products (proteomics), and analyses on human DNA, RNA and chromosomes to detect heritable or acquired disease-related genotypes, mutations, and phenotypes. It includes investigations of how the changes/variations discovered lead to disease processes, monitoring of the effectiveness of chosen precision therapies, and identification of any residual dysfunctions.
Whereas genetic (DNA) testing may inform on anticipated lifetime disease risk, gene expression (RNA) analysis provides a current-state assessment of a disease by looking at the gene expression alterations associated with it. Gene expression levels vary depending on a person’s disease status resulting from genetic and environmental factors interacting over time. As an example of the utility of gene expression (RNA) profiling in molecular pathology and PMed disease diagnostics in cardiology, a brief discussion of the PMed Corus CAD test for hemodynamically significant CAD follows [73,74].
7.1. Peripheral blood gene expression score for obstructive coronary artery disease
Corus® CAD (CardioDx, Inc.) uses age, sex, and the gene expression (RNA) profile of cells derived from a peripheral venous blood specimen (liquid-biopsy) sent under controlled conditions to a certified CardioDx laboratory. The sample is then accessioned, and RNA purification and RNA transcription into cDNA by reverse transcriptase are performed; the cDNA is then used as the template for the quantitative real time PCR. Since mRNA expression profiles can change over several orders of magnitude even in a short time after specimen collection, stabilization using liquid nitrogen, RNase denaturing reagents, or RNA stabilization reagents, is crucial to avoiding inaccurate results.
Corus CAD helps clinicians identify the likelihood that a patient has coronary artery stenosis of at least 50%. It is the only sex-specific test for the evaluation of obstructive CAD because it accounts for cardiovascular differences between men and women [73,74]. It was developed as an alternative to invasive/noninvasive coronary angiography or myocardial perfusion imaging (MPI) by SPECT or PET. It ascertains obstructive CAD in non-diabetic patients with no previously diagnosed myocardial infarction or revascularization, presenting with typical or atypical symptoms suggestive of obstructive CAD. Corus CAD gauges the gene expression (RNA) level of 23 genes to get an individualized score on a scale of 1 to 40—the higher the score, the graver the obstructive disease. The score is used by cardiologists and primary care clinicians for determining whether a non-diabetic patient’s symptoms of cardiovascular disease are due to CAD.
8. Molecular genetic biomarkers
Complex tests, known as biomarker tests for molecularly targeted therapies, have the potential to enable the selection of the most efficacious treatment—and also to signal treatments that may be injurious or ineffectual—for the molecular substructure/underpinnings of an individual patient’s condition. Such tests are key to realizing the promise of PMed. A useful definition encompassing some of the principal uses of a biomarker is “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses” [75].
Humans are genetically polymorphic: no two individuals will have identical nucleotide sequences (apart from identical twins). At any nucleotide locus, some fraction of individuals will carry a different nucleotide than the common one, a phenomenon called Single Nucleotide Polymorphism (SNP, [35]). Molecular genetic biomarkers comprise soluble or suspended molecules—e.g., DNA including SNPs, microRNA, proteins—in the blood, urine, or in secretions. Analysis of the sequences of biomarker DNA, RNA, and proteins enables better understanding of fundamental biological function and interactions. The clinical utility of biomarker tests for molecularly targeted therapies hinges on consistent reliability, a clear-cut clinical objective, and confirmation of improved resultant outcomes. Biomarker test development and application are accelerating at a rapid rate, boosted by new clinical research discoveries yielding improved understanding of the genetic origins of disease. Novel biomarkers may enable the earlier detection of slowly-developing cardiovascular diseases, with more-widespread lifestyle-modifications and therapies for primary prevention.
Biomarkers might be exploited for direct assignment of individual patients to different treatment regimens within a clinical trial (Fig. 3); e.g., enrolling only patients who test positive for a biomarker that is known or presumed to be associated with a higher risk of subsequent events, or with a higher possibility of responding to a specific type of therapy, could reduce the size or duration of a clinical trial.
DNA markers are informative in both basic and applied research, and are useful in the search for functional variants of medically remarkable genes. DNA sequence-based markers as a group may generally affect levels and patterns of gene expression and gene function, and contribute to a person’s susceptibility to various diseases (mutations are one example of changes to DNA sequence). RNA and proteins also contain required information, and therefore demand parallel study. Generally, however, molecular genetic markers have intrinsic limitations. Among limiting factors, genomic and epigenetic heterogeneity can be quite problematic, at times.
8.1. Active mutations' expression may be fluctuating over time or transgenerationally
A one-time single sampling of an individual patient may not reveal the biomarkers needed to determine prognosis and treatment because the molecular signature of disease may actually be shifting/fluctuating over time or remotely in time [76], encompassing transgenerational epigenetic effects [77]. A mutation critical in the early development of a particular disorder may no longer be active, or mutations that are invariably active in one stage may no longer be active in a subsequent one in the natural evolution of the disease. The pace of genome and exome (encompassing all the expressed genes in a genome) sequencing is accelerating, and one way to help identify active mutations could be to use existing sophisticated bioinformatics tools to understand how mutations disrupt networks and to then create comprehensive lists of common and active mutations.
Availability of a wide array of genetic biomarkers offers tools for detection and characterization of genetic variation. Two forms of DNA sequence-based markers, SNPs and simple sequence repeats (SSRs), predominate in contemporary genetic analysis [78,79]. The most studied molecular genetic markers, SNPs, are distributed over the whole genome [80]. The number of SNPs is estimated to fall mostly within a range from 5–10 SNPs/kbp (kilo-base-pairs) [81]. Besides SNPs, there are variations in the number of copies of a DNA sequence in the >1 kbp size-range, which can increase or decrease gene activity levels; they constitute a major source of genetic variation between individuals, underlie many diseases, and are not uncommon in the human genome [82].
9. Pharmacogenomics and pharmacogenetics in personalized medicine
The number of disorders that can be precisely diagnosed and then treated with a highly specific therapy is certain to increase dramatically in the coming years. Today, tests are available to spot many of the genetic differences, allowing drug dosages to be customized. Up until recently, discoveries in molecular genetics had to wait for years to be applied in medical practice and such tests had been underutilized, thus resulting in preventable adverse drug reactions and undoubtedly billions of dollars in avoidable costs.
Currently, pharmacogenomics and pharmacogenetics are getting trendy, due to the promise of targeted medications conjuring images of a future when a test performed by using a simple chip will allow patients to be assigned to PMed treatments that are more efficacious and with fewer, if any, side-effects (cf. Figs 3 and 4). Indeed, in excess of 100 drugs have label information regarding pharmacogenomic biomarkers [83]. Each novel molecular mechanism is now applied promptly after its proper identification. As a result, new emerging molecular-targeted therapies are being regularly introduced into clinical practice.
Aptly responding to the post-genomic era idea that it is feasible to get a molecular diagnosis for individual patients and the ensuing potential for genomic PMed, drug manufacturers have begun amassing enormous troves of human DNA data in hopes of significantly shortening the time it takes to identify new drug candidates. This move is speeding substantially the advent of PMed. Scanning large databases of volunteers, who agree to have their DNA sequenced and to grant access to detailed medical records, is now possible by the plummeting cost of genetic sequencing.
Clinicians should expect that findings of cardiogenetic testing can bring about psychological stress on participants and/or their relatives [84]; for some, WGS will uncover things they were not looking for and might not want to know. And, widespread testing could pose privacy issues because genomic information is digital and would be easy to distribute.
Presently, it remains unanswered whether developing heterogeneous targeted interventions for common multifactorial disorders could become widespread and economically viable. The epigenetic principles clearly suggest that, for complex diseases, DNA will never, on its own, be a powerful divinatory tool, even when gene functioning is well understood. Accordingly, PMed does not restrict itself to genetic or genomics factors but, additionally, integrates environmental or behavioral components. Anyway, considering the by far most commonly distributed in the population multigenic and multifactorial (also encompassing “environmental” factors) diseases, such as CAD, genomic functional prediction in the near future will likely be about as accurate as weather prediction; it will be correct to variable degrees but far from precise. Massive amounts of data and statistical modeling go into a prediction, and stochastic factors can affect the outcome. As data continue to be acquired, it will be critical that the genetics specialists should guide the rest of the medical community through the disruptive technologies and new service models that arise [85,86].
I should now emphasize that PMed is not just about ascertaining most advantageous medications and their dosages. Particularly for some cancers, genetic diagnostic tests can help gauge the aggressiveness of the tumor and, eventually, decide whether, e.g., to perform prostatectomy or apply less invasive treatments. Systems biology approaches that identify master regulatory genes of disease progression may enable the effective stratification of patients—molecular oncologic studies have shown that if a prostate carcinoma lacks genes that produce an aggressive type, the malignant cells may remain stable within the prostate for decades, obviating the need for radical treatments [87].
In the realm of cardiovascular disease, strides are being made toward identifying genetic variants in, e.g., patients who do not respond to particular drugs for managing hypertension and heart failure (ACE inhibitors, beta-blockers, calcium channel blockers, and diuretics). Naturally, after a connection between a gene and a syndrome is ascertained, procuring a reliable diagnostic test for the gene takes time.
9.1. Drug metabolism in individual patients
Most drugs are treated by the body like foreign substances, also known as xenobiotics (from the Gk. xenos “stranger” and biotic “related to living beings”). The drug-metabolizing system of the body, which involves absorption, distribution, biotransformation and excretion, provides a chemical protection generally aimed at detoxifying xenobiotics. Knowledge of pharmacokinetics—how a drug gets into and what happens to it in the body, including its biotransformation/metabolism, in individual patients is key to PMed because it can convey precise understanding of why different patients may not respond in the same way to the same medication (Fig. 4). Numerous distinct enzymes, each corresponding to a different gene/gene-set, regulate how individuals metabolize drugs.
The cytochromes P450s (CYPs, a superfamily of about 50 heme-containing isoenzymes, each encoded by a different gene) are the most important class of drug-metabolizing enzymes in humans; they are located either in the inner membrane of mitochondria or in the endoplasmic reticulum of cells. CYPs metabolize thousands of endogenous and exogenous chemical compounds. Genetic variability (polymorphism) in these enzymes may influence an individual patient’s response to commonly prescribed drug classes [88], including beta blockers. Personalized aberrations in any of these genes can affect both the minimum effective dosage and the maximum one that a person can tolerate without experiencing adverse effect(s). Tissue expression of drug-metabolizing enzymes influences susceptibility to therapeutic and adverse effects of drugs under different conditions (e.g., drug interactions) and in different individuals.
The liver is the major organ of drug metabolism; the extent of metabolism in the major extrahepatic drug-metabolizing organs amounts to approximately 10–20% of the hepatic metabolism. With aging, the liver’s capacity for metabolism through the CYP450 enzyme system is reduced by ≥ 30% because hepatic volume and blood flow are diminished [89]. Thus, drugs that are metabolized through this system reach elevated levels and exhibit prolonged half-lives in the elderly; age-related changes in kidney function can also have similar adverse effects on how fast a drug is eliminated from the body [89].
10. Challenges in implementing personalized medicine
Genetic variation is sizeable, and each individual, with the potential exception of monozygotic twins, possesses a unique DNA sequence. Improved appraisal of genetic diversity in humans requires comprehensive assessments using molecular genetic markers. Genome-wide association studies (GWAS) (see Appendix A.) have begun to identify large numbers of genetic variants that influence the risk of human diseases and variability in high-dimensional human traits/phenotypes where the number of traits assayed per individual can reach many thousand. GWAS search for associations between the genome, typically represented as SNPs, and one phenotypic binary or continuous-valued variable/ trait, such as a disease indicator or biomarker [54]. GWAS have enjoyed increasing popularity in recent years, due primarily to the thousands of genetic variants found to be significantly associated with complex traits [90]; there is currently progress made toward raising the predictive power of polygenic testing through increased GWAS sample sizes and improved genotyping and sequencing technologies.
A striking feature of the newly associated variants is that the top signals often occur at DNA sites that do not encode amino-acids [91] but have more intricate actions. DNA variations encompass polymorphisms ensuing from substitution of single nucleotides (SNPs), insertion or deletion of DNA fragments of different lengths (from a single to several thousands of base pairs), or duplication or inversion of DNA fragments. DNA variations are referred to as “neutral” when they cause no detectable change in metabolic or phenotypic traits; otherwise, they are classified as “functional;” there may be multiple variants of any given gene in the human population, leading to polymorphism.
10.1. Genotyping assays and analysis of pleiotropic polymorphisms
As I noted in discussing the statins earlier, some pleiotropic polymorphisms are neutral, some are damaging and some beneficial, if not immediately, maybe later on in life. Since mutations occur at any place within a gene, there are usually many different alleles for each gene. Some may be neutral and some may be deleterious. It is common in recessive disorders to find that the individual is a compound heterozygote [92]. This means that he/she has two defective genes but they are not mutated at the same site within the gene. Different mutations within the same gene give rise to allelic heterogeneity in the gene pool.
Mutations in key nucleotides of a coding sequence may change the amino-acid composition of a protein, and lead to new molecular conformations and functional variants. Such variants may have an increased or decreased metabolic efficiency compared to the “original,” may lose their functionality completely, or even gain a new/unpredictable function. Mutations in regulatory regions of the genome may affect levels and patterns of gene expression [93]; e.g., turning genes on/off or under/overexpressing proteins in specific tissues at different developmental or subsequent stages of life under various conditions, and may lead to or contribute to a disease diathesis.
In the final analysis, PMed aims at distinguishing categorically between individuals who are more or less likely to respond to a certain medication and, equally, between those who are more or less at risk for any adverse side-effects (Fig. 3). With such information/knowledge, more suitable choices for drug therapies can be formulated to maximize the likelihood of efficacious treatment while minimizing adverse reaction risk (Fig. 4). Progress is indeed being made in this direction: genomics, specific molecular blood biomarker tests, and other pertinent patient characteristics are starting to be applied to predict how an individual with a particular genotype may behave and respond to medications [94,95]. In fact, the genotyping of polymorphic drug-metabolizing enzymes has yielded amended drug dosing for many conditions, such as coronary and peripheral artery disease and their sequelae. This has helped preempt [96,97] dangerous side-effects, harmful drug interactions, or ineffectual treatment (cf. Fig. 2 and Fig. 3).
The genome contains a tremendous amount of data; each individual differs at millions of genetic locations from the reference human genome. Genotyping assays and their analyses are complicated, and proper comprehension of their findings entails a high level of scientific competency. Thus, a conspicuous problem is that the potentially tremendously valuable technology has grown faster than the medical practitioners’ ability to understand the vital results that it generates; an important gap already exists between the rapidly increasing genetic knowledge of cardiac diseases and the medical applications in practice [98].
10.2. Practicality in communicating genomic information in a typical clinical setting
Clinical cardiologists are not molecular geneticists and so a useful test should provide results that can be easily understood by busy clinicians, and must be simple to apply in a typical clinical setting. Gaps in understanding translational genomics, and cross-disciplinary information deficits, along with costs and time-constraints will continue to be challenges to the establishment of an improved pathway of research translation and PMed. On the basic science side, the efficiency of scientific translation can be improved through the use of milestone and outcome-driven management approaches, focusing on research goals and on shortening the time lapse between discovery and development.
Better and quicker approaches are needed for performing economic evaluations of genetic and genomic tests and of the consequences of assaying particular genetic variants. There are significant challenges; because of the large number of bases in the complete haplotype genome, even an accuracy of 99.99 percent will produce >300,000 errors per patient. Furthermore, many different sources of information will need to be brought together efficiently to determine how to move forward with integrating genomic advances into PMed in a way that maximizes patient benefit while at the same time making the most economic sense.
Discovery of genome-enabled biomarkers, construction of longitudinal registries with interconnected molecular and clinical data, design and conduct of biomarker-informed clinical trials, and the advancement of novel practice models and implementation research for the integration of genomic tools in health care systems, including individualized drug development, are all areas that have been attracting attention in recent years. Work in these areas should provide new insights into disease and its treatment. In the end, the success of PMed relies on the collective efforts of clinicians, patients, and researchers to inform research inquiry and translation.
10.3. Conventional clinical trials and innovative PMed/PCard approaches
The preceding considerations require us to examine each of the major points of tangency between the usefulness of conventional clinical trials and the innovative approaches of PMed/PCard in order to determine as best we can whether prevailing conventional population-based trials are confounding knowledge and unduly impinging upon and obstructing PMed/PCard advances. Standard population-based clinical trials gather a handful of measurements from thousands of subjects. At the other extreme, N-of-1 or single subject clinical trials consider an individual patient as the sole unit of observation in a study investigating the predisposition to different interventions and, more specifically, their efficacy or side-effect profiles. PMed/PCard require different ways of testing disease susceptibility and therapeutic interventions. Investigators need to probe the myriad factors—mainly genetic and environmental—that shape a given individual’s susceptibility and response/reaction to a particular treatment.
The question ultimately is whether the cost of specific prospective clinical trials, properly allowing for and characterizing human genome diversity/genomic variances of diverse population subgroups, is too high and, if so, whether it could be offset by the increased effectiveness of a more penetrating/insightful approach in understanding the causes of and determining the treatment of particular human diseases affecting different (genetically definable) patients and groups.
Such multi-group clinical trials will also provide identification/increased knowledge about the factors that lead to disease or to health, and to treatment success or failure. Already, the plethora of genetic data due to technological advances has ushered-in large-scale analyses to determine the molecular underpinnings of human disorders. Sophisticated computational methods have been proposed for predicting which variants are likely to have functional impact and can contribute to changes in phenotype [99,100], and a number of meta-analysis suites exist to annotate variants with the results of the prediction methods [101,102].
People vary in their susceptibility to different diseases, and while much of the explanation will commonly be due to environmental factors like diet, genetic predispositions play a role in many cases. The collection and analysis of DNA samples (see Appendix A) may, in conjunction with epidemiological evidence, help lead to the identification of genetic factors in various human diseases and eventually to ways to treat or prevent those diseases.
I believe most will agree that one or more of several uses of DNA sampling and sequencing justify pursuing this project in PMed/PCard now and with vigor. The definitive answer should be empirically knowable. If in the process there appear to be specific areas in which even modest actions could be taken with the clear expectation of improving pathogenetic understanding and treatment effectiveness, let us take them without undue delay.
11. Future directions for a personalized cardiology: the challenge of the post-genomic era
With sporadic exceptions like blood transfusion—based on the four major blood groups determined by the presence/absence of the A and B antigens on the surface of erythrocytes, and the Rh factor/antigen which can be present or absent (±)—for most of Medicine’s past, the best that physicians could do to cope with disease prevention and treatment was based on the expected response of an average patient. Currently, however, a more personalized/precise approach to medicine is becoming possible, as the cost of sequencing a human genome has declined substantially and is approaching $1000. As a result, we are witnessing an era of precipitous advances in biomedicine and bourgeoning understanding of the genetic basis of cardiovascular and other diseases, reminiscent of the resurgence of innovations in physico-mathematical sciences and biology-anatomy-cardiology in the Renaissance [10,103–106], a parallel time of radical change and reformation of medical knowledge, education and practice.
11.1. Clinical understanding of naturally occurring genomic differences/ variants
Health care providers increasingly have access to unconventional technologies that may identify molecular variations specific to an individual patient, which can subsequently be targeted for treatment. At present, we do not possess the competency to construct biomedicine’s equivalent of the Periodic Table. A complete delineation of the molecular “building blocks” of most multifactorial human disease syndromes (pathology/pathophysiology) still remains sometime into the future. Indeed, the genetic factors contributing to complex trait variation may reside in regulatory (up until recently relegated to “junk DNA” status), rather than protein-coding portions of the genome. Variable penetrance with incomplete expression is common in many cardiac diseases and environmental and modifier gene [35] effects are increasingly appreciated as key components of pronounced genotype-phenotype plasticity.
The current state of understanding of genetics in cardiology is not yet adequate to supplant conventional clinical approaches to diagnosis and therapy to any large extent, but future work will undoubtedly attain much a more precise elucidation of gene variant–environment interactions regulating genotype–disease phenotype plasticity [35].
The current push for meaningful use of electronic health records (EHRs) and health-relevant events/history such as exposures to “environmental” risks, the revelation of the structure of the human genome, gene-gene and gene-environment interactions and the insights that flow from these, and the increasingly broad and pervasive online access to health data/information generally, are all auspicious innovative developments for future advances. Electronic health data include copies of medical histories in textual and spoken form, x-ray and MRI images, and test results from biopsies and genetic microarrays.
Dealing with the data from millions of patients will require computing capabilities beyond those currently being used in medical research, and will require collaborations between medical researchers and a wide range of computer scientists and engineers providing user-friendly software, reliable and secure cloud storage making every synced folder accessible everywhere for viewing or editing files, efficient search algorithms, and stable servers—file sync (synchronization) is a software-supported system keeping up-to-date files stored in different physical locations.
Assembling and managing all of these data is a monumental task, but it could lead to much deeper understanding of disease processes and the way different medications affect different populations. Powerful analyses and tools, tailored to the particular user (e.g., cardiologist-internist-surgeon), will extract useful information from the electronic record, while versatile abstraction and data mining tools will afford concise summaries of the data necessary for a given purpose.
Adaptive learning health systems will use data and technology to be taught by clinical experience. This would be achieved by systematically pulling together various types of patient information/data, including laboratory test results, genomic information, treatments, and clinical outcomes; analyzing the acquired data using artificial intelligence software; and by then translating the knowledge gained from these assessments into clinical practice [107]. Thus supported by multifaceted voluminous data, genomic diagnostics should be able to eventually shed light on the underpinnings of virtually every human malady, because virtually every disease has a genetic component.
If everyone has a whole genome sequence done at birth, it could be used throughout life, and the relevant information could be applied to other family members, as well. As health knowledge is mapped to individuals by the logical aggregation of their lifelong health information, patient-activated, cardiac event monitoring is an economical, diagnostic tool which will allow patients to record arrhythmia or ischemic episodes whenever and wherever they happen with the use of a miniature transmitter and, with the contribution of health professionals, should discern states requiring attention and trigger proactive interventions.
Such comprehensive implementations of genomics in clinical practice can strengthen the evidence-base on genomic approaches to medical practice. Moreover, they will speed up the translation of basic and clinical genomic research findings by encouraging health care professionals and organizations to apply the accruing knowledge to the process of changing routine conventional practice, potentially resulting in improved PMed processes and outcomes. It should be possible to move ahead with implementation/translation even as the evidence database is still expanding. Genomic medicine programs are currently heading in the right direction at several academic medical centers and large integrated health systems [108].
12. Ethical, legal and social implications of incorporating personalized practice into cardiology
Modern molecular genetics/genomics can provide the means to substantially impact many aspects of PCard, including the development of pharmacological treatments. Detailed genetic information enables the grouping of individuals through their polymorphisms. Ensuing studies of, e.g., genetic biomarkers have increased knowledge and aided the prediction of human disease, enabling more accurate diagnoses and improving the safety and efficacy of medications/interventions tailored to the needs of specific patient subgroups. It has also increased our understanding of the molecular mechanisms of diseases, allowing genes and their products to turn into targets of innovative molecular pharmacogenomic analyses and treatments aimed at modulating gene activity. A combination of stem cell technologies, nuclear transfer and epigenetic modulation, plus gene-editing methods has made it generally feasible to manipulate the human genome.
12.1. The RNA-guided CRISPR-Cas9 nuclease system in PCard
Thus, a number of genome editing technologies have emerged, including the RNA-guided CRISPR-Cas9 nuclease system. Originally identified as a natural defense mechanism in bacteria, this system can be easily programed to cut and edit almost any DNA sequence in a variety of cell types and organisms. CRISPR-Cas9 targeted genome editing technology (see Appendix A) holds promise for PMed/PCard, by achieving human genetic modifications allowing targeted changes to almost any gene, and by promoting innovative treatments [109]. Cas9 is a nuclease—enzyme that cleaves the nucleotide chains in nucleic acids into smaller segments—guided by small RNAs through Watson-Crick base pairing with target DNA; it is highly specific, efficient and well-suited for high-throughput genome editing.
Potentially it can cure diseases by disrupting endogenous disease-causing genes, correcting disease causing mutations or inserting new genes with protective functions [110]. Consequently, CRISPR-Cas9 could potentially be used to actually treat complex multigenic cardiovascular disorders, which additionally involve interplay of “environmental” factors, such as coronary artery disease, hypertension, stroke, and hereditary cardiomyopathies, by cutting out defective genes and replacing them with their wild-type versions, or by repairing mutations. However, for the time being this remains theoretical—philosophically inclined readers will note here the Aristotelian potentiality–actuality dichotomy/distinction.
Although such applications augur substantial clinical/health advances, safety concerns and would-be Aristotelian “unintended consequences” render CRISPR-Cas9 genome editing a PMed/PCard topic of ethical debate. We have no knowledge of the reliability or efficiency of human genomic modification in practice. Genes might be more interconnected than we think. Could an intentional alteration in a gene provoke an unintended conversion in another part of the genome, or might it cause even a dysregulation of the entire genome, dispensing catastrophic downstream aftereffects?
Perceptively, Richard Dawkins, the English evolutionary biologist and author, expounds an adroit analogy in several of his books [cf., 111]: the genome not as a blueprint but, rather, as a recipe, or array of instructions for preparing the organism. This is a judicious analogy because, just as there is no way to map one portion of a cake to one part of the recipe, one part of the organism cannot be mapped to one part of the genome; furthermore, an isolated modification in the recipe/genome can modify the cake/organism as a whole—e.g., replacing baking powder with yeast gives a hard, bread-like texture to the cake as a whole.
12.2. Difficult dilemmas posed by genomic screening to identify DNA variants
The use of DNA-based screening for diseases that have a genetic component can pose particularly difficult dilemmas. It is easy to understand why there have been concerns about misuse of genetic information of this type. The prospect that we will be adept at identifying early in life [112], or even “preimplantation” [113], who will manifest serious diseases in middle or even old age raises thorny ethical issues: they pertain to discrimination in health, disability, and life insurance coverage, as well as in employment and, particularly, stigmatization of carrier-status following genetic screening with obstacles in affording/providing medical care [114,115]. The Genetic Information Nondiscrimination Act of 2008 (GINA, [116]) is an Act of Congress in the United States designed to prohibit the use of genetic information in health insurance and employment.
12.3. Disease prediction and prevention
In general, therapeutic genome editing interventions in somatic cells are ethically accepted, allowing for the due balance between risks and benefits and the practice of informed consent. Thus, gene therapy for single-gene recessive disorders requires only gene addition for treatment, provided that the capacity to make sufficient amounts of a functional gene product yields amelioration of the disease. As noted above, often genes have more-than-one/pervading effects, and secondary actions are in most cases unknown, so that altering a large number of genes might incur undetermined hazards. However, genetic manipulation of somatic cells for the purpose of treating or preventing complex cardiovascular diseases poses fewer ethical dilemmas than does manipulation aimed at germ cells or alteration of nondisease genes. Indeed, germline cell manipulations are fraught with danger.
In a pertinent current joint statement seeking a moratorium on the use of gene-editing and gene-altering technologies in the clinical setting, it has been emphasized [117] that “the possibility of human germline engineering has long been a source of excitement and unease among the general public, especially in light of concerns about initiating a ‘slippery slope’ from disease-curing applications toward uses with less compelling or even troubling implications. Manipulations that affect ova, spermatozoa, or totipotent cells such as blastomeres are heritable. Changes made to the DNA in these cells potentially can affect not only the treated individual but also his/her offspring. Thus, CRISPR/Cas9 genome editing can produce (side) effects and unpredictable inheritable changes transmissible to future generations. Clearly, the goals of germline cell manipulations constitute a major focus of the ethical debate that will decide when and if human germline genome engineering is applied [118].
In any case, I suspect that we are a long way away from the time when we will be able to say with corroborated confidence whether a baby will develop such multifactorial diseases in middle life, and when this information will be of any actuarial use. And quality of life for individuals who develop a particular disease is not predictable by genetic tests. Those who have exaggerated the likely speed of scientific progress in this arena, and hence have raised staid ethical concerns about its misapplication, have simply disregarded the enormously complex interactions between nature, nurture, and the pathology of aging, on which most of our common polygenic diseases are based. Further complications are posed by the uncertainties of future options. It is not possible to predict what therapies for management or cure of disease may be developed or when these therapies will be available to patients. Until this field is much further advanced, and adequate pilot studies have been completed to find out how reliable/consistent it may prove in practice, it will be problematic and not yet very productive to elaborate an adequate ethical framework for its wide-scale application.
12.4. The many-sided ramifications of Post-HGP developments that underlie PMed/PCard
There is no doubt that the many-sided ramifications of post-HGP developments in genetics, which underlie PMed/PCard, have had an exceptional impact, provoking much excitement and concerns on the part of both health professionals and the public reflecting on the ethical framework demanded for doctors and the community at large, so as to cautiously contend with this complex and fast moving field. However, there is a critical factor which has hindered rational debate in post-HGP genetics, particularly regarding its PMed/PCard applications. Because its science/data is so new and complex, many doctors and specialists in biomedical ethics have had little more understanding of what is going on than the public at large.
Although a fair number of influential geneticists may have attempted to interpret the field in recorded interviews [119], including social and political issues, for their clinical colleagues and the overall community, potential medical applications/benefits of the post-HGP genetics were mostly communicated by basic scientists who—though “up-to-date” with genetics and molecular biology—had only scanty appreciation as regards the afflictions and concerns of seriously sick people.
In brief, because basic translational and clinical research have progressed so swiftly, there has been a rapidly widening communication gap between the basic biological sciences, the clinical world, bioethicists, and the public. As noted by the philosopher of science Jerome Ravetz, who analyzed scientific knowledge from a social and ethical perspective, science is a “delicate and vulnerable social activity” ([120], p. 72), and it involves many fine value judgments and is guided by the informal controls of scientific leaders and traditions rather than by formal rules and institutions, and to protect their work, scientists came to insist that it be separated from social concerns and that its guiding values all be internal to science itself [120].
However, the fast unfolding developments underlying PCard should be accompanied by broad, pluridisciplinary, collaborative, proactive work to anticipate and address emerging ramifications, including issues of health disparities, bioethics, patient privacy and the physician–patient relationship. To be sure, science is barreling forward, but the ethics aren’t; and we should not expect or want the science to slow down, but we should aspire that the ethics keep pace with it too—to avoid a forthcoming criticism that biomedical ethics are consistently lagging the present.
At the far end of the spectrum of the debate on the post-HGP foundations of PMed/PCard we reenter the “Brave New World” of Aldous Huxley—which he wrote before the structure of DNA was known, and consequently the biological techniques used in it to control the populace do not include genetic manipulations. It conjures fears about the serious misuse of genetics for changing the behavioral patterns of individuals for sinister ends, the augmentation of various physical traits for non-therapeutic purposes (enhance performance of athletes), and the like. Huxley’s novel, of course, is science fiction and such extreme paradigms are so far not based on scientific data. Thus, deliberations on their ethical aspects are not very fruitful. However, placed in a context that recognizes the current state of biomedical scientific progress, they may serve a purpose: undoubtedly, while we are in the midst of a major biomedical scientific/technological revolution, we cannot foretell what will be rendered possible, sooner or later.
12.5. Blending scientific freedom and research-participant/patient dignity
The suitable balance between the values of scientific freedom and dignity of the individual patient/research-participant emerges as the dominant concern of PMed/PCard ethics. This general concern is undergoing a shift to informational and privacy themes in its clinical application. Specific concerns exist about the confidentiality and use of genetic information and results of genetic tests because, unlike many other specimens, genetic liquid biopsy information can be stored for long periods in the form of a frozen blood sample. Such samples provide a source of material that can be analyzed for factors other than that for which it was originally intended, and at a time removed from the collection and consent process.
As PCard becomes incorporated into clinical practice, ethical, legal and social implications should expand in scope and complexity to encompass legal and economic layers added by the potential impact of the patent system, and they will demand growing attention. At the same time, busy clinicians have many more pressing ethical issues to worry them, and for many the ethical concerns of the post-genomic genetics seemingly hold little immediate relevance to day-to-day practice.
12.6. Mushrooming bioinformatics data and inequalities in accessing it
These facts notwithstanding, two broad issues are likely to grow in importance: the repercussions of the mushrooming volume of health information related to PMed/PCard; and concerns regarding the PMed potential to exacerbate disparities in healthcare. Specifically, genomics, proteomics, transcriptomics, metabolomics and other Omics approaches—together with bioinformatics and computational biology technologies that support the integration and processing of genomic records underlying PMed/PCard—yield enormous data compilations. With the HGP, molecular biology became part of the modern trend toward “big data science.” The ensuing application of these prolific technologies in clinical settings depends on sustained and coordinated investments in infrastructure science, such as databases and biobanks, and raises difficult issues related to privacy and discrimination. The availability of such data is also likely to bring about changes in physician–patient relationships and to add to personal injury litigations.
Furthermore, inequalities in research undertakings to assemble biomolecular evidence, as well as in securing access to healthcare services and bioinformatics technologies that make it easier to pull together genomic research data housed in private, public and hybrid “clouds,” are likely to accentuate existing healthcare disparities. Work on these issues will have to focus not only on inequalities in the promises for rational therapeutics and PMed/PCard existing within subpopu-lations in the developed societies, but also those impacting fair access to healthcare in low- and middle-income countries (LMICs) globally [121]. The challenge for innovation in post-genomic PMed/PCard is markedly much greater in LMICs where basic infrastructures are often under-resourced and anticipated institutional relationships can be fragile [122]. It will entail enormous financial costs [123].
12.7. Regulatory agencies’ role in the diffusion of new PMed/PCard treatment modalities
Finally, translation of pharmacogenomics into public health action has been at the epicenter of the biomedical research and development (R&D) agenda since the completion of the HGP. Regulatory agencies nowadays form an important bottleneck before diffusion of new PMed/PCard treatment modalities can take place. Every change in treatment modalities including, e.g., the composition, production, and description of any medication, synthetic biology product, or pharmacogenomics test, and/or their potential use, has to be approved by these agencies, which make the final decisions regarding the indications for which the drug may be prescribed [124].
The agencies also determine a drug’s availability: in hospitals and/or in pharmacies; over the counter or only by prescription; prescribed by general physicians or only by a medical specialist. Additional regulatory agencies/branches aim at assuaging concerns about possible environmental harms, as might be associated with synthetic biology products connected with fundamental genomic studies [125].
13. Conclusions
Genetic variation influences disease predisposition and severity as well as treatment response. The use of genetic information to aid diagnosis, stratify risk and guide therapy can potentially impact most facets of cardiological practice. Now on the horizon is a diverse, individual patient-centered, approach to medical practice that encompasses the development of new, gene-based diagnostics and preventive medicine tactics, and offers the broadest range of personalized therapies based on pharmacogenetics. Development of new technologic approaches is a dynamic ongoing process since technologies themselves, their embedding and their impacts are shaped by complex co-construction processes by the different stakeholders involved. Therefore, the implementation of genomics in PMed/PCard can be accelerated by having ethicists, healthcare industry economists, and regulatory personnel work in-parallel with researchers and clinicians, rather than in-sequence, to effectively and efficiently transform practice.
Advancing knowledge of the genome may fundamentally alter our view of humanity. Over time, translation of genomic and high-tech approaches unquestionably will transform clinical practice in cardiology and medicine as a whole, with the adoption of new PMed procedures. Clearly, future prospects far outweigh present accomplishments, which are best viewed as a promising start. It is now essential for pluridisciplinary health care workers/providers to examine the drivers and barriers to the clinical adoption of the emerging revolutionary paradigm, in order to expedite the realization of its potential. The time has come for all stakeholders to engage with each other in order to learn about relevant societal and ethical aspects of their work, in an effective co-constructive and problem-structuring process to put into operation PMed/PCard by formulating problems that encompass the proper policy objectives, alternatives, and expected outcomes. Ultimately, success must be certified not simply by clinicians, scientists, machines and technicians, but within a wider arena (the agora?) through public engagement. Admittedly, we are not there yet, but we are definitely on our way to achieving the vision of Hippocrates, Osler, and Garrod!
Acknowledgments
I gratefully dedicate this work to the memory of my prized Professors: the patrician Severo Ochoa (1905–1993), 1959 Nobel Prize in Physiology or Medicine for discovering an enzyme that enables the synthesis of RNA; and the unassuming Robert A. Good (1922–2003), 1970 Albert Lasker Clinical Medical Research Award for uniquely important contributions to our understanding of the mechanisms of immunity and for launching the era of cellular engineering. As medical student at New York University School of Medicine (preclinical years), I had the wonders of biochemistry and molecular biology opened up to me by Ochoa; shortly thereafter, Good impressed me at the University of Minnesota Medical School (clinical years) with the urgent need to apply molecular biology to clinical practice for the benefit of individual patients—such as the original “Bubble boy” whom he treated with the first (1968) bone marrow transplants for SCID; sometimes he met with our small student group around his kitchen table, before the crack of dawn!
Acknowledgment of sources of funding
Research support, for work from my Laboratory surveyed here, was provided by: National Heart, Lung, and Blood Institute, Grant R01 NIH 50446; National Science Foundation, Grant CDR 8622201; and North Carolina Supercomputing Center and Cray Research.
Appendix A.
Sequencing technology has exploded since the completion of the HGP. The contemporary sequencing technologies are far faster and cheaper than the prototypical Sanger sequencing, since they proceed in a massively parallel manner (meaning many strands of DNA are sequenced at once), generating far more data per instrument-run than the Sanger method, and thus demanding far bigger data storage facilities plus analysis and manipulation of much larger datasets than in the past. Numerous qualified laboratory exome- (the protein-coding portion of the genome) and genome-sequencing processes are necessary to support the identification, interpretation, and reporting of clinically significant variants.
Targeted approaches employing next-generation sequencing allow researchers to focus time, expenses, and data analysis on specific areas of interest. Such targeted examinations can encompass the exome, particular genes of interest, targets within genes, or mitochondrial DNA. Focused sequencing of a subset of genes or regions of the genome, generates smaller, more manageable data sets, and cuts turnaround times and data analysis costs.
DNA microarrays
To analyze one gene at a time in morphologically preserved tissue sections or cell preparations hybridization methods are applied. Contrariwise, the DNA microarray technology, also called genome chips, or DNA chips, provides a means to simultaneously analyze thousands of the genes. The array is an orderly arrangement of known DNA samples. Microarrays contain thousands of spots of DNA samples, or “probes,” which are commonly less than 200 microns in diameter, and analysis of the microarrays requires special imaging equipment and computer algorithms. Comparing spot portraits among samples of different pathological origins can identify diagnostic or prognostic biomarkers for complex diseases.
The DNA microarray is fabricated by spotting known DNA probes on a solid support such as glass or nylon through the use of high speed robotics [126]. The probes can either be known cDNA sequences (500–5000 bases) or synthetic oligonucleotides (20–80 nucleotides)—cDNA, or complementary DNA is double-stranded DNA synthesized from a single stranded RNA (e.g., messenger RNA (mRNA) or microRNA (microRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. The array is then exposed to fluorescent-labeled cDNA prepared from total sampled mRNA. This free “target” DNA will bind to the spots containing homologous sequences. The identity and abundance of the complementary sequences can then be determined.
It is also possible to mix DNAs prepared with different fluorescent labels. Signals are measured as absolute intensities for a given target, or as ratios of two probes with different fluorescent labels, representing two separate treatments to be compared, or with one probe as an internal control.
Regrettably, arrays suffer from a fundamental “design-bias”—they only return results from those genomic/DNA regions for which probes have been designed. And, as microarrays are limited in the number of single nucleotide polymorphisms (SNPs) they can contain, they tend to focus more on relatively common variants, at the expense of SNPs with lower minor allele frequency (MAF). Accordingly, arrays are only as good as the particular databases from which they are designed (need-based design). Although newer arrays are being designed with lower MAFs in mind, many investigators are drawn toward NGS and higher generation sequencing techniques, as they can pick up both common and rare variants—circumvent the fundamental “design bias” of arrays.
Next (Second)-Generation, and Higher-Generation Sequencing
Next-generation genome sequencing (NGS) [35,127,128] does not have any such design-bias/limitation. It can demonstrate functional DNA sequence variants that influence diseases ranging from atherosclerosis to hypertension, calcific aortic valve disease (CAVD), and familial cardiomyopathies [10,23,24,36,128–131]. NGS technologies are incredibly complex and produce prodigious amounts of sequence data in a massively parallel fashion and relatively inexpensively. They also present a challenge that is beyond technology: how to understand and appropriately translate the biology into something meaningful for individual patients.
Higher- [third- “3G” and fourth-generation “4G”] sequencing techniques are currently becoming available [132–134]. They differ from the NGS technologies, which rely on PCR chain-reaction amplification giving rise to sequencing errors. They can generate rapidly and accurately very long reads of sequence (10–20 kilobases long) from single molecules of DNA. Producing long reads is notable because it is intuitively less complicated to assemble genomes from fewer longer fragments of DNA. While these techniques are still not able to sequence an entire human genome in one run, they possess the potential to become powerful diagnostic tools. One obvious application may be to sequence the smaller genomes of various infectious agents from viral and bacterial outbreaks to identify genetic elements that may result in pathogenicity, survival, or antimicrobial resistance.
Massively parallel RNA sequencing (RNA-Seq)
Unlike DNA, levels of RNA (transcriptome, including mRNA, rRNA, tRNA, and other non-coding RNAs [10,35,128,131] fluctuate in response to the Bernardian environment and other factors. Consequently, sequencing RNA can bring to light an all-encompassing understanding of a person’s state of health at particular successive times. Innovative studies have linked genetic differences between individuals to RNA expression, translation, and protein levels (Fig. 2), although mRNA and protein levels are not mutually correlated that well despite the archetypical paradigm of gene → RNA transcript → protein [35,135, 136].
Regulation of protein levels/function can also occur at any post-transcriptional stage, involving pre-mRNA alternative splicing, the exportation of mature mRNAs from the nucleus, differences in the stability of mRNA molecules in the cytoplasm, factors impinging on the translation process by which polypeptides are constructed from mRNA on ribosomes, polypeptide assembly and 3D folding into functional mature proteins, and post-translational modifications or degradations of the proteins themselves.
Another NGS technology for massively parallel RNA sequencing, known as RNA-Seq [137,138], is emerging as a very effective tool to reveal RNA presence and quantity from a genome of interest at any particular time. In principle, a population of RNA is converted to a library of complementary DNA (cDNA) fragments and is then sequenced in a high-throughput manner [139].
RNA-Seq can be used to analyze the changing cellular transcriptome of bodily organs and tissues, each adapted/specialized to perform distinct functions. Thus, it can show which RNAs are connected with complex diseases and can be used to study quantitative trait loci associated with gene expression in specific syndromes and in response to therapeutics and environmental conditions. RNA-Seq methods cover all aspects of the transcriptome without any a priori knowledge of it, allowing for the analysis of such things as novel transcripts, splice junctions and noncoding RNAs.
Genome-Wide Association Studies (GWAS), Whole Genome Sequencing (WGS), and Whole Exome Sequencing (WES)
Microarray-based genome-wide association studies (GWAS) are often conducted to help identify specific genes underlying complex disorders, and include large numbers of individuals that are screened for polymorphic markers at numerous genetic loci; a statistical association of particular markers with certain disorders indicates that a marked region of the genome could contribute to them. The HapMap Project has exploited GWAS as a powerful tool that facilitates the identification of genetic markers of disease and of genetic associations with complex conditions. For such genotyping studies, microarrays remain widely adopted because they are less expensive than NGS and much more suitable to processing thousands of samples required for typical GWAS. However, a lingering stumbling block is the cost of whole genome sequencing (WGS). To lower the cost per sample and increase the throughput in identifying variants/mutations which may be the root cause of various common human diseases, many investigators are focusing on techniques for sequencing specifically just all of the expressed genes in a genome, i.e., the exome. Whole exome sequencing (WES), currently costs about $400; WES concentrates the sequencing power on <2% (for humans and other mammals) of the genome encompassing the coding sequences for functional proteins and disregarding the rest [140]. Assuming that the most significant mutations occur in coding regions, it becomes efficient, cost-effective, and timesaving to focus attention on exome analysis.
Whole exome sequencing (WES) is sequencing targeted to protein-coding exons and uses thousands of custom probes to extract the <2% of the genome that contains them. WGS entails all-inclusive sequencing of the entire genome; to reduce costs, WGS is typically performed at a lower sequencing depth—this refers to the number of times a base/ nucleotide is read during the sequencing process, and is also called redundancy of coverage [141]—and may lack the capacity to detect low-frequency mutations in heterogeneous samples. WES can yield evidence of exactly how a protein was rendered nonfunctional and why a patient might be susceptible to or actually have a specific disease. To date, demand has existed for tests offering more information, but continued demand will hinge on the cost of the testing, who is paying for it, and its outcomes.
The CRISPR-Cas9 genetic–phenotypic screen platform
Genetic screens are experimental techniques that can identify the causal relationship between genotype and phenotype by introducing perturbations into the genome and/or epigenome with the purpose of evaluating how (epi-) genome aberrations bring about phenotypic variations. Typically, genetic screens are conducted in cultured cells exploiting RNA interference or complementary DNA libraries, which have shortcomings.
Lately, the emergence of methods employing CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats [CRISPR]-associated nuclease 9) introduces a versatile tool for genetic screen studies and genome/epigenome editing [142]. The CRISPR-Cas9 platform, in conjunction with single-gene-knockout collections of human cells [143], covering a sizable and growing fraction of the expressed human genome, allows systematic screens for a wide variety of cellular phenotypes and promises to boost the power of genetic-phenotypic screens in human applications [144].
Remarkable progress has been made over the past dozen years of NGS technology development in terms of speed, nucleic acid residues read-length, and throughput, along with a powerful reduction in per-base cost. These advances have rendered NGS into a molecular microscope and are paving the way for the growth of a large number of novel NGS applications in basic science and in translational research areas impacting PMed, such as clinical diagnostics and pharmacogenetics. Third-generation technologies such as single-molecule sequencing in real time (SMRT) should further overhaul genomics research foundations of PMed, enabling: a more complete characterization of whole genome sequences and the information encoded therein; a more complete characterization of the transcriptome and of epigenetic modification mechanisms involving methylation patterns; and a better appreciation of interactions between proteins and nucleic acids regulating cell processes in development, growth, aging, and disease processes.
References
- 1.van der Eijk PJ. Hippocrates: the protean father of medicine. Lancet. 2002;359(2285) [Google Scholar]
- 2.Sykiotis GP, Kalliolias GD, Papavassiliou AG. Pharmacogenetic principles in the Hippocratic writings. J. Clin. Pharmacol. 2005;45:1218–1220. doi: 10.1177/0091270005281091. [DOI] [PubMed] [Google Scholar]
- 3.Hippocrates. Loeb Classical Library, Hippocrates, Volumes I–VIII. Cambridge, MA: Harvard University Press [The name of each treatise is given in brackets in the text citation]; [Google Scholar]
- 4.Pontius JU, Mullikin JC, Smith DR, Agencourt Sequencing Team. Lindblad-Toh K, Gnerre S. et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiter LT, Potocki L, Chien S, Gribskov M, Bier EA. systematic analysis of human disease-associated gene sequences in Drosophila melanogaster . Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien S, Reiter LT, Bier E, Gribskov M. Homophila: human disease gene cognates in Drosophila . Nucleic Acids Res. 2002;30:149–151. doi: 10.1093/nar/30.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttmacher AE, Collins FS, Drazen JM, editors. Genomic Medicine: Articles from the New England Journal of Medicine. Baltimore: Johns Hopkins University Press; 2004. [Google Scholar]
- 8.Shim Y, Hampton TG, Straley CA, Harrison JK, Spero LA, Bashore TM, Pasipoularides AD. Ejection load changes in aortic stenosis. Observations made after balloon aortic valvuloplasty. Circ. Res. 1992;71:1174–1184. doi: 10.1161/01.res.71.5.1174. [DOI] [PubMed] [Google Scholar]
- 9.Jameson JL, Longo DL. Precision medicine—personalized, problematic, and promising. N. Engl. J. Med. 2015;372:2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 10.Pasipoularides A. Heart’s Vortex: Intracardiac Blood Flow Phenomena. Shelton, CT: People’s Medical Publishing House; 2010. [Google Scholar]
- 11.Pasipoularides A, Murgo JP, Bird JJ, Craig WE. Fluid dynamics of aortic stenosis: mechanisms for the presence of subvalvular pressure gradients. Am J Physiol: Heart Circ Physiol. 1984;246:H542–H550. doi: 10.1152/ajpheart.1984.246.4.H542. [DOI] [PubMed] [Google Scholar]
- 12.Pasipoularides AD, Shu M, Womack MS, Shah A, von Ramm O, Glower DD. RV functional imaging:3-D echo-derived dynamic geometry andflow field simulations. Am. J. Phys. Heart Circ. Phys. 2003;284:H56–H65. doi: 10.1152/ajpheart.00577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasipoularides A, Shu M, Shah A, Womack MS, Glower DD. Diastolic right ventricular filling vortex in normal and volume overload states. Am. J. Physiol. Heart Circ. Physiol. 2003;284:H1064–H1072. doi: 10.1152/ajpheart.00804.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasipoularides A, Vlachos PP, Little WC. Vortex formation time is not an index of ventricular function. J. Cardiovasc. Transl. Res. 2015;8:54–58. doi: 10.1007/s12265-015-9607-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasipoularides A, Shu M, Shah A, Tucconi A, Glower DD. RV instantaneous intraventricular diastolic pressure and velocity distributions in normal and volume overload awake dog disease models. Am. J. Physiol. Heart Circ. Physiol. 2003;285:H1956–H1965. doi: 10.1152/ajpheart.00372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasipoularides A. Clinical assessment of ventricular ejection dynamics with and without outflow obstruction. J. Am. Coll. Cardiol. 1990;15:859–882. doi: 10.1016/0735-1097(90)90287-y. [DOI] [PubMed] [Google Scholar]
- 17.Isaaz K, Pasipoularides A. Noninvasive assessment of intrinsic ventricular load dynamics in dilated cardiomyopathy. J. Am. Coll. Cardiol. 1991;17:112–121. doi: 10.1016/0735-1097(91)90712-i. [DOI] [PubMed] [Google Scholar]
- 18.Mirsky I, Pasipoularides A. Clinical assessment of diastolic function. Prog. Cardiovasc. Dis. 1990;32:291–318. doi: 10.1016/0033-0620(90)90018-w. [DOI] [PubMed] [Google Scholar]
- 19.Georgiadis JG, Wang M, Pasipoularides A. Computational fluid dynamics of left ventricular ejection. Ann. Biomed. Eng. 1992;20:81–97. doi: 10.1007/BF02368507. [DOI] [PubMed] [Google Scholar]
- 20.Hampton T, Shim Y, Straley C, Pasipoularides A. Finite element analysis of cardiac ejection dynamics: ultrasonic implications, Am Soc Mechan Engin. Bioengin Div. (ASME BED) Adv Bioengin. 1992;22:371–374. [Google Scholar]
- 21.Hampton T, Shim Y, Straley C, Uppal R, Smith PK, Glower D, Pasipoularides A. Computational fluid dynamics of ventricular ejection on the CRAY Y-MP. IEEE Proc Comp Cardiol. 1992;19:295–298. [Google Scholar]
- 22.Pasipoularides A. Cardiac mechanics: basic and clinical contemporary research. Ann. Biomed. Eng. 1992;20:3–17. doi: 10.1007/BF02368503. [DOI] [PubMed] [Google Scholar]
- 23.Pasipoularides A. Fluid dynamic aspects of ejection in hypertrophic cardiomyopathy. Hell. J. Cardiol. 2011;52:416–426. [PMC free article] [PubMed] [Google Scholar]
- 24.Pasipoularides A. LV twisting and untwisting in HCM: ejection begets filling. Diastolic functional aspects of HCM. Am. Heart J. 2011;162:798–810. doi: 10.1016/j.ahj.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasipoularides A. Invited Comment: functional imaging (FI) combines imaging datasets and computational fluid dynamics to simulate cardiac flows. J. Appl. Physiol. 2008;105:1015–1016. doi: 10.1152/japplphysiol.zdg-8134-vpcomm.2008. [DOI] [PubMed] [Google Scholar]
- 26.Pasipoularides A, Palacios I, Frist W, Rosenthal S, Newell JB, Powell WJ., Jr Contribution of activation-inactivation dynamics to the impairment of relaxation in hypoxic cat papillary muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1985;248:R54–R62. doi: 10.1152/ajpregu.1985.248.1.R54. [DOI] [PubMed] [Google Scholar]
- 27.Pasipoularides A, Murgo JP, Miller JW, Craig WE. Nonobstructive left ventricular ejection pressure gradients in man. Circ. Res. 1987;61:220–227. doi: 10.1161/01.res.61.2.220. [DOI] [PubMed] [Google Scholar]
- 28.Pasipoularides A. Complementarity and competitiveness of the intrinsic and extrinsic components of the total ventricular load: demonstration after valve replacement in aortic stenosis. Am. Heart J. 2007;153:4–6. doi: 10.1016/j.ahj.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasipoularides A, Mirsky I, Hess OM, Grimm J, Krayenbuehl HP. Myocardial relaxation and passive diastolic properties in man. Circulation. 1986;74:991–1001. doi: 10.1161/01.cir.74.5.991. [DOI] [PubMed] [Google Scholar]
- 30.Pasipoularides A. Right and left ventricular diastolic flow field: why are measured intraventricular pressure gradients small? Rev. Esp. Cardiol. 2013;66:337–341. doi: 10.1016/j.rec.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasipoularides A. Right and left ventricular diastolic pressure-volume relations: a comprehensive review. J. Cardiovasc. Transl. Res. 2013;6:239–252. doi: 10.1007/s12265-012-9424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasipoularides A. Evaluation of right and left ventricular diastolic filling. J. Cardiovasc. Transl. Res. 2013;6:623–639. doi: 10.1007/s12265-013-9461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasipoularides AD, Shu M, Shah A, Glower DD. Right ventricular diastolic relaxation in conscious dog models of pressure overload, volume overload, and ischemia. J. Thorac. Cardiovasc. Surg. 2002;124:964–972. doi: 10.1067/mtc.2002.126677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasipoularides A. Fluid dynamics of ventricular filling in heart failure: overlooked problems of RV/LV chamber dilatation. Hell. J. Cardiol. 2015;56:85–95. [PMC free article] [PubMed] [Google Scholar]
- 35.Pasipoularides A. Linking genes to cardiovascular diseases: gene action and gene-environment interactions. J. Cardiovasc. Transl. Res. 2015;8:506–527. doi: 10.1007/s12265-015-9658-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasipoularides A. Calcific aortic valve disease: Part 1 - molecular pathogenetic aspects, hemodynamics and adaptive feedbacks. J. Cardiovasc. Transl. Res. 2016;9:102–118. doi: 10.1007/s12265-016-9679-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Priori SG, Napolitano C, Memmi M, et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002;106:69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 38.Nava A, Bauce B, Basso C, et al. Clinical profile and long-term follow-up of 37 families with arrhythmogenic right ventricular cardiomyopathy. J. Am. Coll. Cardiol. 2000;36:2226–2233. doi: 10.1016/s0735-1097(00)00997-9. [DOI] [PubMed] [Google Scholar]
- 39.L Hedley P, Jørgensen P, Schlamowitz S, Moolman-Smook J, Kanters JK, A Corfield V, Christiansen M. The genetic basis of Brugada syndrome: a mutation update. Hum. Mutat. 2009;30:1256–1266. doi: 10.1002/humu.21066. [DOI] [PubMed] [Google Scholar]
- 40.Arvind P, Jayashree S, Jambunathan S, Nair J, Kakkar VV. Understanding gene expression in coronary artery disease through global profiling, network analysis and independent validation of key candidate genes. J. Genet. 2015;94:601–610. doi: 10.1007/s12041-015-0548-3. [DOI] [PubMed] [Google Scholar]
- 41.Winkelmann BR, Hager J, Kraus WE, et al. Genetics of coronary heart disease: current knowledge and research principles. Am. Heart J. 2000;140:S11–S26. doi: 10.1067/mhj.2000.109636. [DOI] [PubMed] [Google Scholar]
- 42.Schunkert H, Erdmann J, Samani NJ. Genetics of myocardial infarction: a progress report. Eur. Heart J. 2010;31:918–925. doi: 10.1093/eurheartj/ehq038. [DOI] [PubMed] [Google Scholar]
- 43.Linsel-Nitschke P, Samani NJ, Schunkert H. Sorting out cholesterol and coronary artery disease. N. Engl. J. Med. 2010;363:2462–2463. doi: 10.1056/NEJMcibr1010765. [DOI] [PubMed] [Google Scholar]
- 44.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J. Am. Coll. Cardiol. 2012;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 45.Lopes LR, Rahman MS, Elliott PMA. systematic review and meta-analysis of genotype-phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart. 2013;99:1800–1811. doi: 10.1136/heartjnl-2013-303939. [DOI] [PubMed] [Google Scholar]
- 46.Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH. Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era. J. Mol. Diagn. 2013;15:158–170. doi: 10.1016/j.jmoldx.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Rau CD, Lusis AJ, Wang Y. Genetics of common forms of heart failure: challenges and potential solutions. Curr. Opin. Cardiol. 2015;30:222–227. doi: 10.1097/HCO.0000000000000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skrzynia C, Berg JS, Willis MS, Jensen BC. Genetics and heart failure: a concise guide for the clinician. Curr. Cardiol. Rev. 2015;11:10–17. doi: 10.2174/1573403X09666131117170446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X, Feng T, Tayo BO, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am. J. Hum. Genet. 2015;96:21–36. doi: 10.1016/j.ajhg.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato N, Loh M, Takeuchi F, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehret GB, Caulfield MJ. Genes for blood pressure: an opportunity to understand hypertension. Eur. Heart J. 2013;34:951–961. doi: 10.1093/eurheartj/ehs455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams R. Joseph Goldstein and Michael Brown: demoting egos, promoting success. Circ. Res. 2010;106:1006–1010. doi: 10.1161/RES.0b013e3181dcab4d. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein JL, Brown MS. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnett DK. Genetics of CVD in 2015: Using genomic approaches to identify CVD-causing variants. Nature Reviews Cardiology. 2016;13:72–74. doi: 10.1038/nrcardio.2015.202. [DOI] [PubMed] [Google Scholar]
- 55.Baker M. Functional genomics: the changes that count. Nature. 2012;482:257–262. doi: 10.1038/482257a. [DOI] [PubMed] [Google Scholar]
- 56.L Hall J. Translating genetic discoveries to improvements in cardiovascular care: the path to personalized medicine. J. Cardiovasc. Transl. Res. 2008;1:37–40. doi: 10.1007/s12265-008-9010-8. [DOI] [PubMed] [Google Scholar]
- 57.Cambien F, Tiret L. Genetics of cardiovascular diseases: from single mutations to the whole genome. Circulation. 2007;116:1714–1724. doi: 10.1161/CIRCULATIONAHA.106.661751. [DOI] [PubMed] [Google Scholar]
- 58.Menasha JD, Schechter C, Willner J. Genetic testing: a physician’s perspective. Mt Sinai J. Med. 2000;67:144–151. [PubMed] [Google Scholar]
- 59.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol. Med. 2001;7:201–204. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- 60.Mukherjee D, Topol EJ. Pharmacogenomics in cardiovascular diseases. Prog. Cardiovasc. Dis. 2002;44:479–498. doi: 10.1053/pcad.2002.123467. [DOI] [PubMed] [Google Scholar]
- 61.Ridker PM, MacFadyen JG, Fonseca FA, et al. Number needed to treat with rosuvastatin to prevent first cardiovascular events and death among men and women with low low-density lipoprotein cholesterol and elevated high-sensitivity C-reactive protein: justification for the use of statins in prevention: an intervention trial evaluating rosuvastatin (JUPITER) Circ Cardiovasc. Qual. Outcomes. 2009;2:616–623. doi: 10.1161/CIRCOUTCOMES.109.848473. [DOI] [PubMed] [Google Scholar]
- 62.Zhou Q, Liao JK. Pleiotropic effects of statins-basic research and clinical perspectives. Circ. J. 2010;74:818–826. doi: 10.1253/circj.cj-10-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao JK, Laufs U. Pleiotropic effects of statins. Annu. Rev. Pharmacol. Toxicol. 2005;45:89–118. doi: 10.1146/annurev.pharmtox.45.120403.095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rocca B, Petrucci G. Personalized medicine, pharmacogenetics, and clopidogrel: unraveling variability of response. Mol. Interv. 2010;10:12–19. doi: 10.1124/mi.10.1.4. [DOI] [PubMed] [Google Scholar]
- 65.Holmes DR, Jr, Dehmer GJ, Kaul S, et al. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”. Circulation. 2010;122:537–557. doi: 10.1161/CIR.0b013e3181ee08ed. [DOI] [PubMed] [Google Scholar]
- 66.Dangas G, Mehran R, Guagliumi G, et al. Role of clopidogrel loading dose in patients with ST-segment elevation myocardial infarction undergoing primary angioplasty: results from the HORIZONS-AMI (harmonizing outcomes with revascularization and stents in acute myocardial infarction) trial. J. Am. Coll. Cardiol. 2009;54:1438–1446. doi: 10.1016/j.jacc.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 67.Lobo I. Same genetic mutation, different genetic disease phenotype. Nature Education. 2008;1(1):64. http://www.nature.com/scitable/topicpage/same-genetic-mutation-different-genetic-disease-phenotype-938. [Google Scholar]
- 68.De Backer J. Cardiovascular characteristics in Marfan syndrome and their relation to the genotype. Verh. K. Acad. Geneeskd. Belg. 2009;71:335–371. [PubMed] [Google Scholar]
- 69.Peluffo AE. The “Genetic Program”: behind the genesis of an influential metaphor. Genetics. 2015;200:685–696. doi: 10.1534/genetics.115.178418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto M, Wakatsuki T, Hada A, Ryo A. Use of serial analysis of gene expression (SAGE) technology. J. Immunol. Methods. 2001;250:45–66. doi: 10.1016/s0022-1759(01)00305-2. [DOI] [PubMed] [Google Scholar]
- 71.Gowda GA, Zhang S, Gu H, Asiago V, Shanaiah N, Raftery D. Metabolomics-based methods for early disease diagnostics. Expert. Rev. Mol. Diagn. 2008;8:617–633. doi: 10.1586/14737159.8.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenberg S, Elashoff MR, Beineke P, et al. For the PREDICT (Personalized Risk Evaluation and Diagnosis in the Coronary Tree) Investigators. Multicenter validation of the diagnostic accuracy of a blood-based gene expression test for assessing obstructive coronary artery disease in nondiabetic patients. Ann. Intern. Med. 2010;153:425–434. doi: 10.7326/0003-4819-153-7-201010050-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ladapo JA, Budoff M, Ross L, et al. Primary endpoint results from a community-based registry evaluating the use of a blood-based age/sex/gene expression test in patients presenting with symptoms suggestive of obstructive coronary artery disease: the PRESET registry. Circ Cardiovasc. Qual. Outcomes. 2015;8:A142. [Google Scholar]
- 74.Lansky A, Elashoff MR, Ng V, et al. A gender-specific blood-based gene expression score for assessing obstructive coronary artery disease in nondiabetic patients: results of the Personalized Risk Evaluation and Diagnosis in the Coronary Tree (PREDICT) Trial. Am. Heart J. 2012;164:320–326. doi: 10.1016/j.ahj.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 75.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 76.van Abeelen AF, Elias SG, Bossuyt PM, et al. Famine exposure in the young and the risk of type 2 diabetes in adulthood. Diabetes. 2012;61:2255–2260. doi: 10.2337/db11-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanson MA, Skinner MK. Developmental origins of epigenetic transgenerational inheritance. Environ. Epigenet. 2016;2:1–9. doi: 10.1093/eep/dvw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burnside RD, Pappas JG, Sacharow S, et al. Three cases of isolated terminal deletion of chromosome 8p without heart defects presenting with a mild phenotype. Am J. Med. Genet. 2013;A161A:822–828. doi: 10.1002/ajmg.a.35699. [DOI] [PubMed] [Google Scholar]
- 79.Phillips III JA, Poling JS, Phillips CA, et al. Synergistic heterozygosity for TGFbeta1 SNPs and BMPR2 mutations modulates the age at diagnosis and penetrance of familial pulmonary arterial hypertension. Genet Med. 2008;10:359–365. doi: 10.1097/GIM.0b013e318172dcdf. [DOI] [PubMed] [Google Scholar]
- 80.Scheinfeldt LB, Tishkoff SA. Recent human adaptation: genomic approaches, interpretation and insights. Nat Rev. Genet. 2013;14:692–702. doi: 10.1038/nrg3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang B, Canestaro WJ, Choudhry NK. Clinical evidence supporting pharmacogenomic biomarker testing provided in US Food and Drug Administration Drug Labels. JAMA Intern. Med. 2014;174:1938–1944. doi: 10.1001/jamainternmed.2014.5266. [DOI] [PubMed] [Google Scholar]
- 84.Caleshu C, Kasparian NA, Edwards KS, Yeates L, Semsarian C, et al. Interdisciplinary psychosocial care for families with inherited cardiovascular diseases. Trends Cardiovasc. Med. 2016;26:647–653. doi: 10.1016/j.tcm.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 85.Katsanis SH, Katsanis N. Molecular genetic testing and the future of clinical genomics. Nat. Rev. Genet. 2013;14:415–426. doi: 10.1038/nrg3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grody WW, Thompson BH, Hudgins L. Whole-exome/genome sequencing and genomics. Pediatrics. 2013;132(Suppl. 3):S211–S215. doi: 10.1542/peds.2013-1032E. [DOI] [PubMed] [Google Scholar]
- 87.Shen M, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma Q. L.A.Y Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol. Rev. 2011;63:437–459. doi: 10.1124/pr.110.003533. [DOI] [PubMed] [Google Scholar]
- 89.Klotz U. Pharmacokinetics and drug metabolism in the elderly. Drug Metab. Rev. 2009;41:67–76. doi: 10.1080/03602530902722679. [DOI] [PubMed] [Google Scholar]
- 90.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res. 2010;20:311–319. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kamphans T, Sabri P, Zhu N, et al. Filtering for compound heterozygous sequence variants in non-consanguineous pedigrees. PLoS ONE. 2013;8:70151. doi: 10.1371/journal.pone.0070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jaffe AS, Babuin L, Apple FS. Biomarkers in acute cardiac disease: the present and the future. J. Am Coll. Cardiol. 2006;48:1–11. doi: 10.1016/j.jacc.2006.02.056. [DOI] [PubMed] [Google Scholar]
- 95.Jesse RL. On the relative value of an assay versus that of a test: a history of troponin for the diagnosis of myocardial infarction. J. Am. Coll. Cardiol. 2010;55:2125–2128. doi: 10.1016/j.jacc.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 96.Sim SC, Kacevska M, Ingelman-Sundberg M. Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J. 2013;13:1–11. doi: 10.1038/tpj.2012.45. [DOI] [PubMed] [Google Scholar]
- 97.Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charron P. Clinical genetics in cardiology. Heart. 2006;92:1172–1176. doi: 10.1136/hrt.2005.071308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Capriotti E, Calabrese R, Fariselli P, Martelli PL, Altman RB, Casadio R. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics. 2013;14(Suppl. 3):S6. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 101.Wang J, Ronaghi M, Chong SS, Lee CG. pfSNP: an integrated potentially functional SNP resource that facilitates hypotheses generation through knowledge syntheses. Hum. Mutat. 2011;32:19–24. doi: 10.1002/humu.21331. [DOI] [PubMed] [Google Scholar]
- 102.Capriotti E, Altman RB, Bromberg Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics. 2013;14(Suppl. 3):S2. doi: 10.1186/1471-2164-14-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pasipoularides A. Galen, father of systematic medicine. An essay on the evolution of modern medicine and cardiology. Int. J. Cardiol. 2014;172:47–58. doi: 10.1016/j.ijcard.2013.12.166. [DOI] [PubMed] [Google Scholar]
- 104.Pasipoularides A. Historical continuity in the methodology of modern medical science: Leonardo leads the way. Int. J. Cardiol. 2014;171:103–115. doi: 10.1016/j.ijcard.2013.11.133. [DOI] [PubMed] [Google Scholar]
- 105.Pasipoularides A. Greek underpinnings to his methodology in unraveling De Motu Cordis and what Harvey has to teach us still today. Int. J. Cardiol. 2013;168:3173–3182. doi: 10.1016/j.ijcard.2013.07.253. [DOI] [PubMed] [Google Scholar]
- 106.Pasipoularides A. Historical Perspective: Harvey’s epoch-making discovery of the Circulation, its historical antecedents, and some initial consequences on medical practice. J. Appl. Physiol. 2013;114:1493–1503. doi: 10.1152/japplphysiol.00216.2013. [DOI] [PubMed] [Google Scholar]
- 107.Krumholz HM. Big data and new knowledge in medicine: the thinking, training, and tools needed for a learning health system. Health Aff. 2014;33:1163–1170. doi: 10.1377/hlthaff.2014.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet. Med. 2013;15:258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 110.Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos. Trans. R. Soc. B. 2016;371 doi: 10.1098/rstb.2015.0496. http://dx.doi.org/10.1098/rstb.2015.0496 pii: 20150496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dawkins R. The Blind Watchmaker. New York: Penguin Books; 2006. pp. 295–297. [Google Scholar]
- 112.American Academy of Pediatrics. Ethical and policy issues in genetic testing and screening of children. Pediatrics. 2013;131:620–622. doi: 10.1542/peds.2012-3680. [DOI] [PubMed] [Google Scholar]
- 113.Ethics Committee of the American Society for Reproductive Medicine. Use of preimplantation genetic diagnosis for serious adult onset conditions: a committee opinion. Fertil. Steril. 2013;100:54–57. doi: 10.1016/j.fertnstert.2013.02.043. [DOI] [PubMed] [Google Scholar]
- 114.Klitzman R. Views of discrimination among individuals confronting genetic disease. J. Genet. Couns. 2010;19:68–83. doi: 10.1007/s10897-009-9262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rioux PP. Clinical trials in pharmacogenetics and pharmacogenomics: methods and applications. Am. J. Health Syst. Pharm. 2000;57:887–898. doi: 10.1093/ajhp/57.9.887. [DOI] [PubMed] [Google Scholar]
- 116.Laedtke AL, O’Neill SM, Rubinstein WS, Vogel KJ. Family physicians’ awareness and knowledge of the Genetic Information Nondiscrimination Act (GINA) J. Genet. Couns. 2012;21:345–352. doi: 10.1007/s10897-011-9405-6. [DOI] [PubMed] [Google Scholar]
- 117.Baltimore D, Berg P, Botchan M, et al. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348:36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bosley KS, Botchan M, Bredenoord AL, et al. CRISPR germline engineering-the community speaks. Nat. Biotechnol. 2015;33:478–486. doi: 10.1038/nbt.3227. [DOI] [PubMed] [Google Scholar]
- 119.Harper PS. Recorded interviews with human and medical geneticists. Hum. Genet. 2016 Nov 15; doi: 10.1007/s00439-016-1744-9. http://dx.doi.org/10.1007/s00439-016-1744-9 [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 120.Ravetz JR. Scientific Knowledge and Its Social Problems. Oxford: Oxford Univ. Press; 1971. [Google Scholar]
- 121.Özdemir V, Muljono DH, Pang T, et al. Asia-Pacific Health 2020 and Genomics without Borders: co-production of knowledge by science and society partnership for global personalized medicine. Curr. Pharmacogenomics Pers. Med. 2011;9:1–5. doi: 10.2174/187569211794728841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huzair F, Borda-Rodriguez A, Upton M. Twenty-first century vaccinomics innovation systems: capacity building in the global south and the role of product development partnerships (PDPS) OMICS. 2011;15:539–543. doi: 10.1089/omi.2011.0036. [DOI] [PubMed] [Google Scholar]
- 123.Hather GJ, Haynes W, Higdon R, et al. The United States of America and scientific research. PLoS ONE. 2010;5(8):e12203. doi: 10.1371/journal.pone.0012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bijker WE, Hughes TP, Pinch TJ, editors. The Social Construction of Technological Systems: New Directions in the Sociology and History of Technology. Cambridge, MA: MIT Press; 1987. [Google Scholar]
- 125.Yearley S. Cultures of Environmentalism: Empirical Studies in Environmental Sociology. Basingstoke, UK: Palgrave Macmillan; 2005. [Google Scholar]
- 126.Hoyt PR, Tack L, Jones BH, Van Dinther J, Staat S, Doktycz MJ. Automated high-throughput probe production for DNA microarray analysis. Biotechniques. 2003;34:402–407. doi: 10.2144/03342mt05. [DOI] [PubMed] [Google Scholar]
- 127.Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 128.Pasipoularides A. Mechanotransduction mechanisms for intraventricular diastolic vortex forces and myocardial deformations: Part 2. J. Cardiovasc. Transl. Res. 2015;8:293–318. doi: 10.1007/s12265-015-9630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pasipoularides A. Calcific aortic valve disease: Part 2 – morphomechanical abnormalities, gene reexpression and gender-effects on ventricular hypertrophy and its reversibility. J. Cardiovasc. Transl. Res. 2016;9:374–399. doi: 10.1007/s12265-016-9695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pasipoularides A. Diastolic filling vortex forces and cardiac adaptations: probing the epigenetic nexus. Hell. J. Cardiol. 2012;53:458–469. [PMC free article] [PubMed] [Google Scholar]
- 131.Pasipoularides A. Mechanotransduction mechanisms for intraventricular diastolic vortex forces and myocardial deformations: Part 1. J. Cardiovasc. Transl. Res. 2015;8:76–87. doi: 10.1007/s12265-015-9611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ku C-S, Roukos DH. From next-generation sequencing to nanopore sequencing technology: paving the way to personalized genomic medicine. Expert Rev. Med. Devices. 2013;10:1–6. doi: 10.1586/erd.12.63. [DOI] [PubMed] [Google Scholar]
- 133.Bleidorn C. Third generation sequencing: technology and its potential impact on evolutionary biodiversity research. Syst. Biodivers. 2016;14:1–8. [Google Scholar]
- 134.Pennisi E. Genome sequencing. Search for pore-fection. Science. 2012;336:534–537. doi: 10.1126/science.336.6081.534. [DOI] [PubMed] [Google Scholar]
- 135.Cenik C, Cenik ES, Byeon GW, et al. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 2015;25:1610–1621. doi: 10.1101/gr.193342.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu L, Candille SI, Choi Y, et al. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Costa V, Aprile M, Esposito R, Ciccodicola A. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur. J. Hum. Genet. 2013;21:134–142. doi: 10.1038/ejhg.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Costa V, Angelini C, De Feis I, Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq. J. Biomed. Biotechnol. 2010;2010:853916. doi: 10.1155/2010/853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 141.Sims D, Sudbery I, Ilott N, et al. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 142.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bürckstümmer T, Banning C, Hainzl P, et al. A reversible gene trap collection empowers haploid genetics in human cells. Nat. Methods. 2013;10:965–971. doi: 10.1038/nmeth.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xue HY, Ji LJ, Gao AM, et al. CRISPR-Cas9 for medical genetic screens: applications and future perspectives. J. Med. Genet. 2016;53:91–97. doi: 10.1136/jmedgenet-2015-103409. [DOI] [PubMed] [Google Scholar]