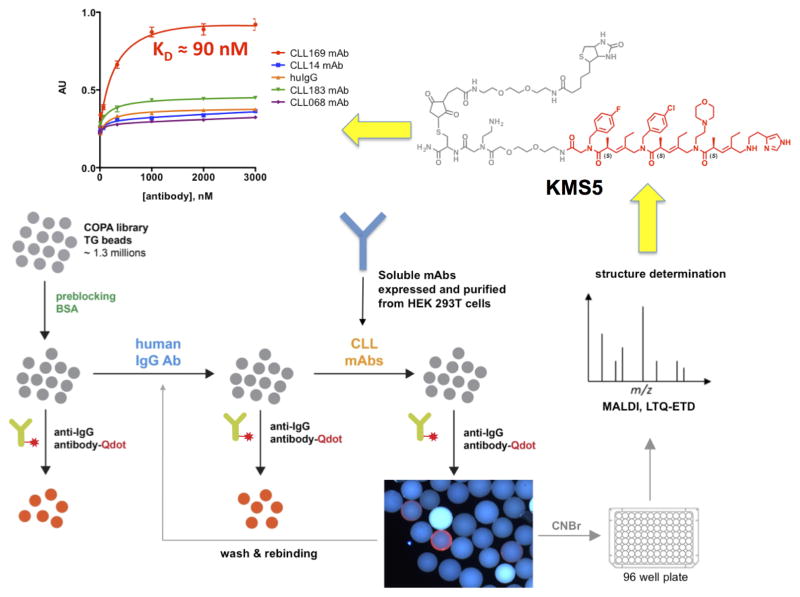
Figure 4. Workflow for the discovery of “epitope surrogates”.
An OBOC library (gray spheres) of over one million compounds comprised of a peptoid unit followed by three COPA21 (chiral oligomer of pentenoic amide) units was first cleared of ligands to uninteresting antibodies. This was done by incubation with a mixture of human IgGs from healthy donors followed, after washing, by addition of a quantum dot-conjugated secondary antibody. After another wash, beads that displayed a high level of fluorescence under a low power fluorescence microscope were removed from the library. The remainder of the bead population was then mixed with soluble versions of the CLL BCR targets. Beads displaying ligands for these species were picked using the same fluorescence-guided method. 16 strongly fluorescent beads were picked. The compounds were released from the beads and sequenced by mass spectrometry. After resynthesis with a new linker including a biotin tag (grey atoms in the chemical structure), the molecules were immobilized on an ELISA plate and binding of the CLL antibodies or control molecules was evaluated (top left graph). The best ligand for the CLL BCR 169 was KMS5 (shown in red), which bound to CLL 169 with a KD of 90 nM. Modified from ref. 22.