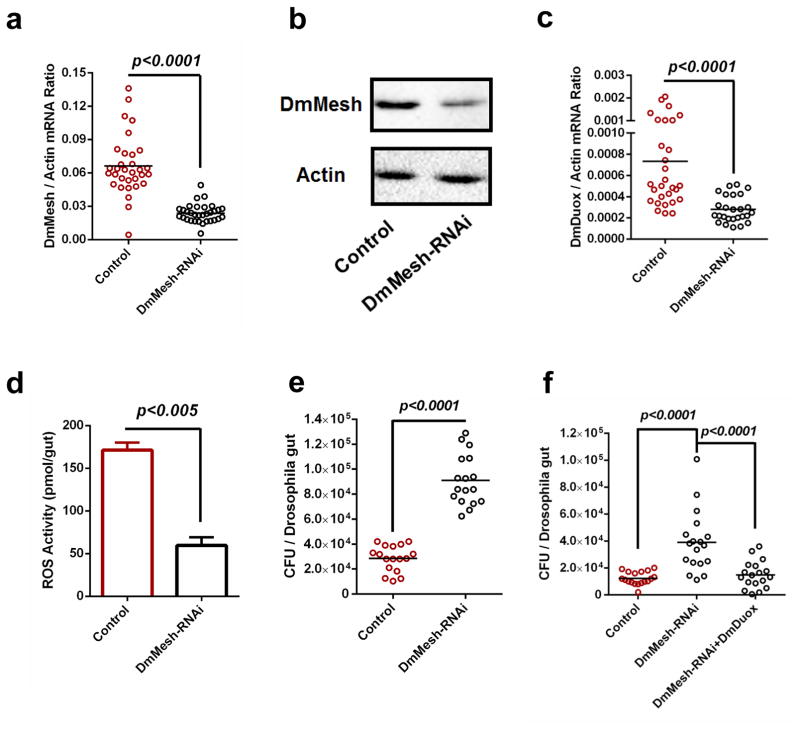
Figure 2. Regulation of commensal microbiome and DmDuox expression in the guts of DmMesh RNAi Drosophila.
(a–b) Knockdown efficiency of DmMesh gene in the guts of DmMesh RNAi Drosophila. The DmMesh RNAi Drosophila strain was generated using a GAL4 line driven by a midgut-specific NP3084 promoter. The NP3084/GFP-RNAi flies served as a negative control. The knockdown efficiency in the guts of DmMesh RNAi flies was assessed via SYBR Green qPCR (a) and immuno-blotting with an AaMesh antibody (b). (b) A total of 50 μg of protein from mosquito lysates was loaded into each lane.
(c–d) Regulation of the DmDuox gene (c) and ROS activity (d) in the guts of DmMesh RNAi Drosophila. (d) ROS activity was measured by H2O2 assay. The data are presented as the mean ± S.E.M.
(e) Regulation of the gut microbiome in the guts of DmMesh RNAi Drosophila. The burden of the gut microbiome was determined by a colony-forming unit (CFU) assay.
(f) Reduction of burden of gut microbiome by rescuing DmDuox into the DmMesh RNAi flies. The burden of the gut microbiome was determined by a CFU assay.
(a, c) The gene expression was determined by SYBR Green qPCR and normalized against D. melanogaster actin (CG12051). The qPCR primers are described in Supplementary Table 6. One dot represents one fly gut. The horizontal line represents the mean value of the results.
(a–f) All results were repeated in at least 3 independent experiments.