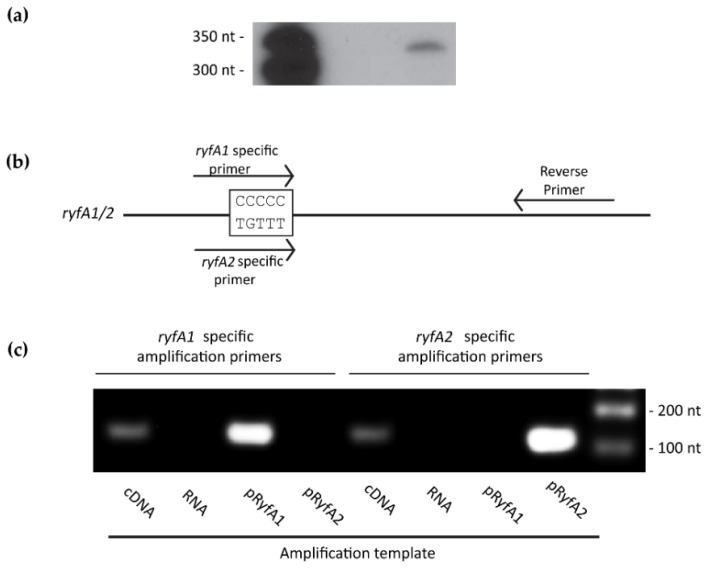
Figure 3.
S. dysenteriae produces both RyA1 and RyfA2. (a) Northern blot analysis using total RNA isolated from wild-type S. dysenteriae following growth to the mid-logarithmic phase at 37 °C and a radio-labeled probe specific to sequences conserved between RyfA1 and RyfA2. The predicted sizes of RyfA1 and RyfA2 are 303 and 305 nt, respectively. (b) Schematic depicting the location and sequence specificity of the amplification primers used in the reverse transcriptase analysis. Each forward primer overlaps the five-nucleotide variable region of RyfA1 or RyfA2, thus providing specificity of amplification. (c) Reverse transcriptase PCR demonstrating that both RyfA1 and RyfA2 are produced by wild-type S. dysenteriae under the conditions tested. Using the ryfA1 specific forward primer, amplification occurs when complementary DNA (cDNA) generated from S. dysenteriae or a plasmid carrying ryfA1 is used as a template (pRyfA1), but not when a plasmid carrying ryfA2 is used as a template (pRyfA2). Similarly, using the ryfA2 specific forward primer, amplification occurs when cDNA generated from S. dysenteriae or a plasmid carrying ryfA2 is used as a template, but not when a plasmid carrying ryfA2 is used as a template. The RNA used to generate the cDNA amplification template is used itself as a template with each primer set to ensure that the sample is free from DNA contamination.