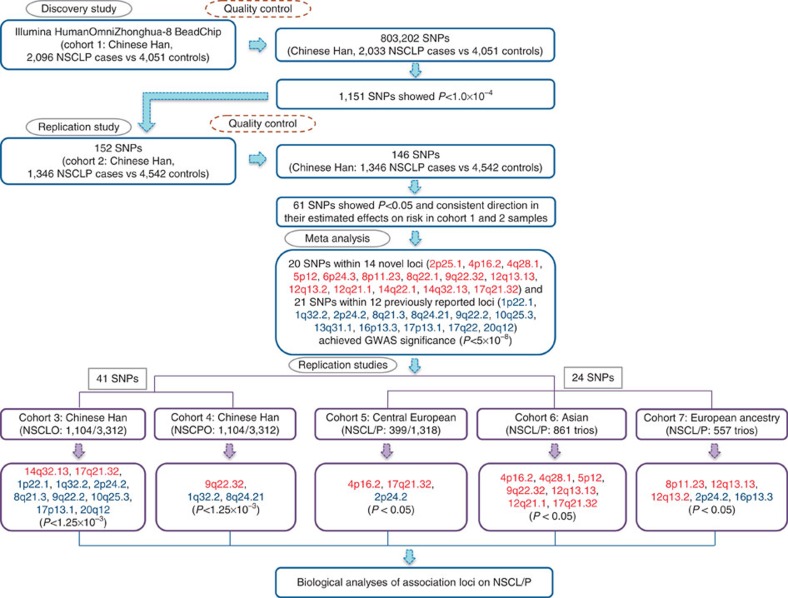
Figure 1. Study design.
We first conducted a GWAS study in 2,096 Chinese NSCLP cases and matched 4,051 controls using Illumina HumanOmniZhonghua-8 BeadChip. After quality control, 803,202 SNPs were remained and analysed in 2,033 NSCLP cases and 4,051 controls, and 1,151 SNPs showed P<1.0 × 10−4 using logistic regression in the discovery stage. One hundred and fifty-two SNPs with P<1.0 × 10−4 were selected for replication in an independent Chinese cohort including 1,346 NSCLP cases and 4,542 controls. After quality control, 146 SNPs remained, of which 61 SNPs showed P<0.05 using logistic regression and consistent direction in their estimated effects on risk in the discovery and validation samples. Then, a fixed-effects meta-analysis of the combined cohorts 1 and 2 samples identified 14 novel loci (20 SNPs) and confirmed 12 previously reported ones (21 SNPs) associated at genome-wide significance (Pmeta<5 × 10−8 using Cochran–Mantel–Haenszel test). We genotyped 41 top SNPs in further 1,104 NSCLO, 1,104 NSCPO patients and 3,312 shared controls in Chinese Han population, respectively. As a result, ten and three loci showed significant associations in cohort 3 and 4 samples (PBonferroni<1.25 × 10−3 using logistic regression and Bonferroni correction). The 24 SNPs (20 from the 14 novel loci and 4 from two newly reported NSCL/P loci) out of the 41 SNPs were also evaluated in Central European, Asian and European ancestry populations, and 3, 7 and 5 loci showed evidence of association in the different cohorts, respectively (P<0.05 using logistic regression). We additionally explored the molecular functionalities of risk variants and their related genes using several complementary methods.