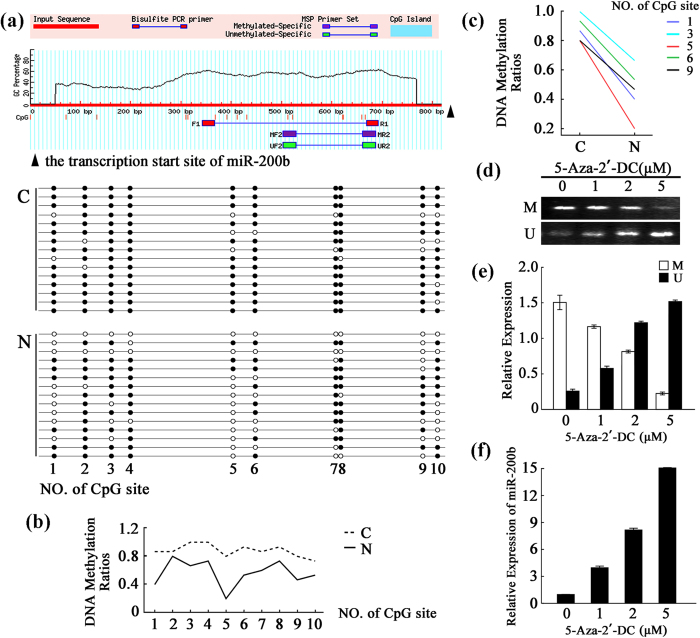
Figure 5. Up-regulation of miR-200b was associated with hypomethylation of DNA in the promoter region in nicotine-treated VSMCs.
(a) CpG methylation in the control and nicotine-treated VSMCs. CpG islands were predicted and bisulfite-sequencing PCR primers were designed using the Methprimer tool. The CpG islands were tinted blue and the BSP primers flanked a 350-bp PCR product upstream of the transcription start site of miR-200b. Each spot indicates one methylation site (CpG); the black spots indicate methylated cytosines, and the white spots indicate unmethylated cytosines. (b) A line graph of the DNA methylation ratios in CpG sites 1–10 from control and nicotine-treated VSMCs. (c) A histogram of the DNA methylation ratios in CpG sites 1, 3, 5, 6 and 9 in control and nicotine-treated VSMCs. (d–f) Verification of DNA methylation regulation in VSMCs. The panels (d) show the MSP results for the primers that were designed for the methylated and unmethylated sites in VSMCs that were treated with a gradient of 5-Aza-2′-DC concentrations. The results are shown in histogram (e). Histogram f shows the results of the real-time PCR analysis of the miR-200b expression level in VSMCs treated with 5-Aza-2′-DC. The values are expressed as the mean ± SEM (n = 5).