Abstract
Clinical electroencephalography (EEG) is the most important investigation in the diagnosis and management of epilepsies. An EEG records the electrical activity along the scalp and measures spontaneous electrical activity of the brain. Because the EEG signal is complex, its interpretation is known to produce moderate inter-observer agreement among neurologists. This problem can be addressed by providing clinical experts with the ability to automatically retrieve similar EEG signals and EEG reports through a patient cohort retrieval system operating on a vast archive of EEG data. In this paper, we present a multi-modal EEG patient cohort retrieval system called MERCuRY which leverages the heterogeneous nature of EEG data by processing both the clinical narratives from EEG reports as well as the raw electrode potentials derived from the recorded EEG signal data. At the core of MERCuRY is a novel multimodal clinical indexing scheme which relies on EEG data representations obtained through deep learning. The index is used by two clinical relevance models that we have generated for identifying patient cohorts satisfying the inclusion and exclusion criteria expressed in natural language queries. Evaluations of the MERCuRY system measured the relevance of the patient cohorts, obtaining MAP scores of 69.87% and a NDCG of 83.21%.
Introduction
Clinical electroencephalography (EEG) is an electrophysiological monitoring method used to record electrical activity of the brain. Clinical EEG is the most important investigation in the diagnosis and management of epilepsies and can also be used to evaluate other types of brain disorders1. An EEG records the electrical activity along the scalp and measures spontaneous electrical activity of the brain. Unfortunately, as noted in by Beniczky et al.2, the EEG signal is complex and inter-observer agreement for EEG interpretation is known to be moderate. Such interpretations of EEG recordings are available in EEG reports. As more clinical EEG data becomes available, the interpretation of EEG signals can be improved by providing neurologists with results of search for patients that exhibit similar EEG characteristics. Searching the EEG signals and reports results in the identification of patient cohorts that inform the clinical decision of neurologists and enable comparative clinical effectiveness research3. For example, a neurologist suspecting that her patient has epilepsy potential formulated the query (Q1) History of seizures and EEG with TIRDA without sharps, spikes, or electrographic seizures. When inspecting the EEG signals and reports of the resulting patient cohort, the neurologist was able to observe the specific features of the EEG for patients that exhibited epileptic potential. In another search instance, a neurologist researcher was interested in one of the research priorities for improving surveillance and prevention of epilepsy as reported by England et al.4, namely, to identify effective interventions for epilepsy accompanied by mental health comorbidities. This researcher formulated the query (Q2) History of Alzheimer and abnormal EEG. The patient cohort that was identified enabled the researcher to observe the treatment outcomes as well as the clinical correlations documented in the EEG reports.
To ensure that patients from a cohort satisfy the criteria expressed in the natural language queries formulated by neurologists, it is important to not only consider the narrative from EEG reports, but also the EEG signal data. Searching for patient cohorts by considering EEG signals and reports relies on (1) an index of the EEG clinical information and (2) relevance models that identify records of relevant patients against a query. Indexing EEG clinical information requires organizing both narratives from the EEG reports and signal data from the EEG recordings. Consequently, the EEG index needs to capture multi-modal clinical knowledge processed both from the reports and the signal recordings. While medical language processing enables the indexing of information form the EEG reports, the index must also comprise a representation of EEG signal recordings. In addition, the relevance models used by the patient cohort retrieval system must account for inclusion and exclusion criteria inferred from processing the natural language query. To address these problems, we have developed a patient cohort retrieval system which produces a multi-modal index of big EEG data. We have also implemented two relevance models to identify the most relevant patients based on their EEG reports and also based on the properties of the EEG signal recordings. The patient cohort retrieval system, called MERCuRY (Multi-modal EncephalogRam patient Cohort discoveRY), uses medical language processing to identify the inclusion and exclusion criteria from the queries and to index the clinical knowledge from the EEG reports. In addition, MERCuRY has two novel aspects not present in previous approaches for patient cohort retrieval: (1) it uses deep learning to represent the EEG signal and to produce a multi-modal EEG index; and (2) it operates based on two EEG relevance models – one that uses only the clinical information from the EEG reports, and a second one which also considers the EEG signal information. We evaluated MERCuRY by using expert judgements of queries against a collection of nearly 20,000 EEGs.
Background
The ability to automatically identify patient cohorts satisfying a wide range of criteria – including clinical, demographic, and social information – has applications in numerous use cases, as pointed out in by Shivade et al.5 including (a) clinical trial recruitment; (b) outcome prediction; and (c) survival analysis. Although the identification of patient cohorts is a complex task, many systems aiming to resolve it automatically have used statistical techniques or machine learning methods taking advantage of natural language processing (NLP) of the clinical documents5. However, these systems cannot rank the identified patients based on the relevance of the patient to the cohort criteria. This notion of relevance is at the core of information retrieval (IR) systems. Thus, viewing the problem of patient cohort identification as an IR problem enables us to not only identify which patients belong to a cohort, but to also rank patients based on relevance to the inclusion and exclusion criteria used in the query.
Using information retrieval for patient cohort identification was considered in 20116 and 20127 by the Medical Records track in the annual Text REtrial Conference (TREC) hosted by the National Institute for Standards and Technology (NIST). When patient cohort identification systems are presented with a query expressing the inclusion/exclusion criteria for a desired patient cohort, a ranked list of patients representing the cohort is produced where each patient may be associated with multiple medical records. Thus, identifying a ranked list of patients is equivalent to producing a ranked list of sets of medical records, each pertaining to a patient belonging to the cohort. In MERCuRY, we have adopted the same framework of identifying patients that are relevant to a cohort and ranking them according to their relevance to the given cohort criteria. However, unlike the TREC patient cohort retrieval systems, which considered only the clinical texts available from a large set of electronic medical records, MERCuRY uses a multi-modal index that encodes textual data available from EEG reports as well as signal data produced by EEG signal recordings.
Historically, the majority of multi-modal retrieval systems have operated on text and image data. For example, Demner-Fushman et al.8 designed a biomedical article retrieval system which allows users to not only discover biomedical articles relevant to a query, but to also discover similar images to those found in any retrieved articles. Their approach clusters images using a large number of visual features capturing color, edge, texture, and other image information. By contrast, our approach relies on unsupervised deep learning to generate a fingerprint of EEG data. As such, although designed for EEG data, our architecture can be easily adapted to support other types of (physiological) waveform data (such as ECGs). Other forms of physiological waveform data were previously investigated by the AALIM9 system which enabled cardiac decision support by allowing physicians to locate similar patients according to the ECG, echo, and audio data associated with the patient. However, unlike our approach, their system does not support search: it can only identify similar patients to provided ECG, echo, or audio recording and thus cannot be used to discover patients matching arbitrary criteria.
The MERCuRY system presented in this paper relies on medical language processing techniques that were informed by the experience gained from participating in the 2010, 2011 and 2012 Informatics for Integrating Biology and the Bedside (i2b2) Challenges on NLP for Clinical Records, which focused on the automatic identification of medical concepts and events in clinical texts10. In contrast, the EpiDEA patient cohort identification system11 which also retrieves EEG-specific patient cohorts, operates on discharge summaries to recognize patients that are relevant only to some pre-defined queries obtained by relying on the EpSO ontology12.
Data
The MERCuRY system was developed to identify patient cohorts from the big EEG data available from the Temple University Hospital (TUH) EEG Corpus13 (over 25,000 sessions and 15,000 patients collected over 12 years). This dataset is unique because, in addition to the raw signal information, physician’s EEG reports are provided for each EEG. Following the American Clinical Neurophysiology Society Guidelines for writing EEG reports14, the EEG reports from the TUH corpus start with a CLINICAL HISTORY of the patient, describing the patient’s age, gender, and relevant medical conditions at the time of the recording (e.g., “after cardiac arrest”) followed by a list of the medications which may influence the EEG. The INTRODUCTION section is the depiction of the techniques used for the EEG (e.g. “digital video EEG”, “using standard 10-20 system of electrode placement with 1 channel of EKG”), as well as the patient’s conditions prevalent at the time of the recording (e.g., fasting, sleep deprivation) and level of consciousness (e.g. “comatose”). The DESCRIPTION section is the mandatory part of the EEG report, and it provides a description of any notable epileptiform activity (e.g. “sharp wave”), patterns (e.g. “burst suppression pattern”) and events (“very quick jerks of the head”). In the impression section, the physician states whether the EEG readings are normal or abnormal. If abnormal, then the contributing epileptiform phenomena are listed. The final section of the EEG report, the CLINICAL CORRELATIONS section explains what the EEG findings mean in terms of clinical interpretation15 (e.g. “very worrisome prognostic features”). Each EEG report in the TUH corpus is associated with the EEG signal recording it interprets. The signal information consists of 24 to 36 channels of signal data as well as an additional annotation channel providing markers identifying events of interest to physicians and technicians. EEG signals are sampled at a rate of 250 Hz or 256 Hz using 16-bits per sample. Each EEG recording from the TUH EEG corpus contains roughly 20 megabytes of raw data, stored in the European Data Format (EDF+) file schema16.
Methods
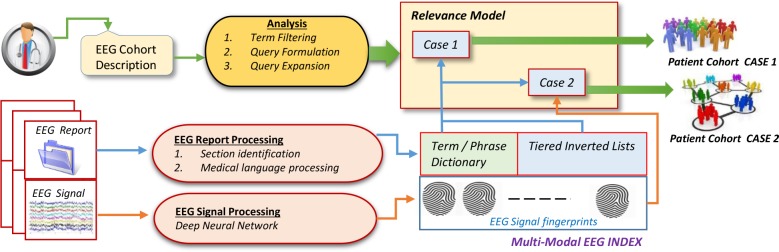
MERCuRY is a multi-modal patient cohort discovery system which allows neurologists to inspect the EEG records as well as the EEG signal recordings of patients deemed relevant to a query expressing inclusion and exclusion criteria through natural language. As illustrated in Figure 1, the neurologist query is analyzed to identify the inclusion and exclusion criteria. The results of query analysis inform two different relevance models (illustrated as Case 1 and Case 2 in Figure 1) which rely on the multi-modal EEG index encoding information identified in the EEG reports and signal recordings. When EEG reports are indexed, the sections of the EEG reports are identified and medical language processing is performed to identify the terms and phrases of the dictionary and to create tiered inverted lists. When the EEG signal recordings are processed, they are represented by EEG signal fingerprints which are produced by deep learning methods. EEG signal recordings are converted into low-dimensional fingerprint vectors which are included in the multi-modal index. Additional details of the index are provided later in the paper. As shown in Figure 1, in MERCuRY we considered two relevance models designed to identify and rank patients based on their relevance to the patient cohort query: Case 1, in which the EEG signal fingerprints are ignored and only the EEG reports are used, and Case 2 in which both the EEG fingerprints and reports are used. These two cases allowed us to experimentally evaluate the impact of the EEG signal fingerprint representation on the overall performance of MERCuRY in identifying patient cohorts.
Figure 1:
Overview of the MERCuRY Patient Cohort Discovery System
A. Indexing the EEG Big Data
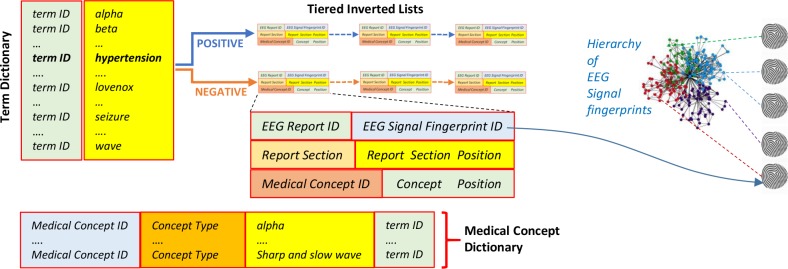
The multi-modal index used by the MERCURY system organizes the information from the EEG reports as well as the information from the EEG signal recordings. It contains both a term dictionary and a medical concept dictionary, listing all the terms and medical concepts discerned from the EEG reports. We considered five medical concept types: (1) medical problems, (2) medical tests; (3) medical treatments (including medications); (4) EEG patterns and activities; as well as (5) EEG events. Because medical concepts often are multi-term expressions (e.g. “spike and slow waves”), the medical concept dictionary used term IDs to associate a concept with all terms expressing it (e.g. “spike and slow waves” is associated with the terms “spike”, “slow” and “wave”). Moreover, as illustrated in Figure 2, each entry from the term dictionary is linked to a pair of inverted lists: the first corresponding to positive polarity associated with the term while the second corresponding to negative polarity. By using polarity information (which is automatically processed from the medical language used in the EEG reports), we have designed a multi-tiered index. Each of the tiered inverted lists is implemented as a linked list. Each cell of those lists indicates for every occurrence of the term: (1) in which EEG report the term was observed; (2) the EEG signal fingerprint of that EEG report; (3) in which section of the EEG report was the term observed; (4) in which position of the EEG section; (5) whether the term belongs to a medical concept identified in the EEG report; and (6) if so, what position does the term have in the concept.
Figure 2:
The Multi-Modal Tiered Index used in the MERCuRY Patient Cohort Discovery System
The EEG signal fingerprints are representations of the EEG signal recordings obtained through deep-learning techniques (described later in the paper) and organized in a similarity-based hierarchy which enables the discovery of relevant patients when the EEG signal recordings are also considered (Case 2 illustrated in Figure 1). When the EEG signal recordings are not used for identifying patient cohorts (Case 1 illustrated in Figure 1) only the term dictionary, the medical concept dictionary, and the tiered inverted lists from the index are used. Creating the multi-modal tiered index for the MERCuRY Patient Cohort Discovery System involves (1) the recognition of sections of the EEG reports; (2) medical language processing to determine (a) the terms from the dictionary, (b) their polarity and (c) the medical concepts; (3) generating the fingerprints for the EEG recording; and (4) organizing the EEG signal fingerprints in the similarity-based hierarchy.
Section Identification. Sections were identified through a rule-based section segmentation approach. Our rules were defined after manually reviewing 300 randomly sampled EEG reports. We detected a set of candidate headers by discovering all sequences of all capitalized words ending in a colon or line break, and normalized section titles based on simple regular expressions. For example, “description of the record”, “description of record”, and “description of the recording” would all be normalized to DESCRIPTION.
Medical Language Processing. In order to build (1) the term dictionary; (2) the medical concept dictionary and (3) the two polarity-informed posting lists we have used the following sequence of steps:
(Step 1) Tokenizing the EEG reports: we relied on Stanford’s CoreNLP pipeline17 to detect sentences and tokens from every EEG report.
(Step 2) Discovering the Dictionary Terms: Each token was normalized in order to account for any lexical variation (e.g. “waves” and “wave” or “markedly and “marked”) using Stanford’s CoreNLP lemmatizer17. The resultant lemmatized terms formed the basis of the dictionary.
(Step 3) Identifying the Polarity: Term polarity was cast as a classification problem implemented as a conditional random field18 (CRF). Leveraging our previous experience with the i2b2 challenge, the CRF assigned a binary polarity value (i.e. positive or negative) to each term based on feature vector containing lexical information as well as information from external resources, such as the NegEx negation detection system19, the Harvard General Inquirer20, the Unified Medical Language System (UMLS) meta-thesaurus21, and MetaMap22. Specifically, we considered nine features: (1) the section name, (2) whether the term was considered a modifier by NegEx, (3) whether the term was within a NegEx negation span, (4) whether the term was in the ‘IF’ category of the Harvard General Inquirer, (5) the part-of-speech tag assigned to the token by Stanford’s CoreNLP part-of-speech tagger17, (6) whether the term belonged to a UMLS concept, (7) whether the term belongs to a MetaMap concept, (8) the original cased term before lemmatization, and (9) the lowercased and lemmatized form of the term. The classifier was trained using 2,349 manual annotations.
(Step 4) Identifying Medical Concepts: An automatic system for medical concept recognition previously developed for the 2010 i2b2 challenge23 recognized medical problems, tests, treatments. In addition, we have produced 4,254 new annotations for EEG patterns and events as well as EEG activities and re-trained the concept recognizer to identify all these types of concepts. Concept extraction was cast as a classification task, in which a CRF was used to detect medical concept boundaries. A support vector machine24 (SVM) was used to classify each concept into one of five types: medical problem, medical test, medical treatment, EEG activity, or EEG event.
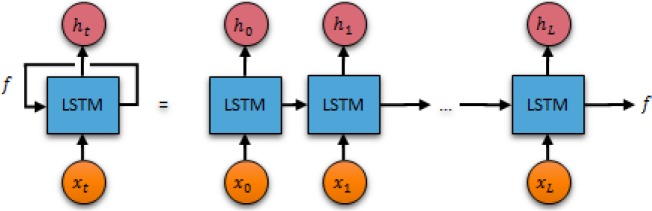
Generating Fingerprints of EEG Signal Recordings. In the TUH EEG corpus, the EEG signals are encoded as dense floating-point matrices of the form D ∈ ℝN×L, where N ∈ [24,36] is the number of electrode potential channels in the EEG and L is the number of samples (such that duration of the EEG recording in seconds is equal to L / 250 Hz). Thus Dij encodes the magnitude of the potential recording on the i-th channel during the j-th time sample. Both the number of channels and the number of samples vary not only across patients, but also across EEG recording sessions. These variations, particularly when considered with the large amount of data encoded in each matrix (typically 20 megabytes), make it difficult to not only characterize the relevance EEG signals to a particular patient cohort, but also to determine the similarity between two EEG signals. For example, consider that a single naïve pass over the TUH EEG corpus requires considering over 400 gigabytes worth of information. Consequently, we devised a representation of the EEG recordings that not only requires less memory, but enables rapid similarity detection between EEG signal recordings. This allows us to compactly encode the information from the EEG signals (reducing 20 megabytes of signal data to a few hundred bytes). Our representation is based on EEG signal recording fingerprints obtained with a recurrent neural network. Recurrent neural networks are deep learning architectures which enabled us to generate fingerprints for each EEG in the TUH EEG corpus in a matter of hours instead of weeks. Because traditional neural networks have difficulty operating on sequential data (e.g. EEG signals) as they cannot consider relationships between successive inputs (e.g. between successive samples), we have used a recurrent neural network25 (a neural network with a loop) which allowed information learned at each sample to persist between samples in the same EEG signal. The learned information (known as the internal memory state) is updated with each sample until ultimately becoming the fingerprint for that EEG recording. Figure 3 illustrates the recurrent neural network used for EEG fingerprinting. As shown, the unrolled network (on the right) processes each sample from the EEG signal and predicts the value of the next sample (ht+1) according to both the current sample (xt) and the current fingerprint (f).
Figure 3:
The recursive neural network used for generating EEG signal fingerprints.
Recursive neural networks are able to connect information from the previous sample to the current sample, allowing them to consider the structure of the EEG waves in each channel. However, interpreting EEGs requires considering not just the immediately adjacent signal information but also long distance signal patterns (e.g. alpha waves being interrupted by a sharp and slow wave complex or repeated bursts of high amplitude delta waves). In order to allow our EEG fingerprints to consider this type of long-distance information – that is, to consider the context of the entire signal when predicting the next sample, we adapt a special form of recursive neural network cell – the Long Short Term Memory26 (LSTM) cell – which are able to remember information for long periods of time.
The recursive neural network shown in Figure 3 is formally defined as follows. We define the parameter K as the fingerprint dimensionality, or the number of dimensions of the finger-print vector such that K ≪ N × L (where N is the number of channels and L is the number of samples). This allows us to determine the fingerprint vector f ∈ ℝ1×K for an EEG D ∈ ℝN×L by using the fingerprint as the internal memory state of the LSTM chain. In order to ensure that the fingerprint can be used to reconstruct portions of the EEG data, we define an additional parameter, W, the sample window, which indicates the number of subsequent samples which should be predicted from each cell. This allows us to learn the optimal fingerprint for each document by determining the vector f which minimizes the cosine distance between the predicted values for each sample (hi, hi+1, …, hi+w) and the actual values (xi+1, xi+2, …, xi+ W+1). Note, that the fingerprint vector and prediction vector are both K-dimensional, while each sample vector is only N-dimensional. Thus, before comparing, we must project the output vector hi into N dimensions by defining a projection matrix W ∈ ℝK×N and a bias vector b ∈ ℝ1×N. Unlike the fingerprint vectors, which are optimized for each individual EEG, W and b are optimized over the entire corpus. Thus, the optimal fingerprint vector f for each document was computed by minimizing the cosine distance between the output of the LSTM cell and the next W samples:
where refers to the standard LSTM loss function26. As defined, the recurrent neural network allows us to generate the optimal fingerprint f for each EEG signal by discovering the vector f which is best able to predict the progression of samples in each EEG recording according to the LSTM. In this way, the fingerprint is able to provide a substantially compressed view of the EEG signal while still retaining many of the long-term interactions and characteristics of the signal.
Organizing EEG fingerprints in a similarity-based Hierarchy. Rapid computation of similarity between EEG signals (or their fingerprints) is facilitated by the Fast Library for Approximate Nearest Neighbors27 (FLANN). FLANN provides implementations of a variety of highly-efficient structures for computing (or approximating) the nearest neighbors of vectors in high dimensions. This allowed us to not only compactly store the EEG signal information, but also to retrieve, for any EEG fingerprint, the most similar EEGs (i.e. the nearest neighbors) as measured by cosine distance. We used a k-means tree which allows for high precision retrieval of the nearest EEGs to any fingerprint by recursively clustering the fingerprint vectors using k-means clustering. The number of clusters is determined by FLANN’s auto-tuning mechanism.
B. Query Analysis
The purpose of query analysis is to identify the inclusion and exclusion criteria expressed in the query. For example, in the query “patients with shifting arrhythmic delta suspected of underlying cerebrovascular disease” two separate inclusion criteria are detected: “shifting arrhythmic delta” and “cerebrovascular disease”. Similarly, in the query “patients with dementia and no abnormal EEG”, there is one inclusion criterion, namely “dementia” and one exclusion criterion, namely “abnormal EEG”. To detect automatically the criteria, the following steps are used:
(Step 1) Term Filtering: Tokenization, lemmatization, and part-of-speech tagging using Stanford’s CoreNLP pipeline17 enables the filtering of terms that are not identified as a noun, verb, adverb, adjective, or preposition.
(Step 2) Query Formulation: Our approach for determining inclusion and exclusion criteria in the query relied on the same polarity and medical concept classifiers used (and previously described) for building the inverted index from EEG reports. Specifically, we considered two methods for recognizing inclusion and exclusion criteria: (a) phrase chunking using Stanford’s CoreNLP pipeline and (b) medical concept detection using the previously described classifier. In both cases, we relied on the previously described polarity classifier to distinguish between inclusion criteria (positive) and exclusion criteria (negative) based on the polarity of each phrase or concept.
(Step 3) Query Expansion: In order to account for the fact that many medical concepts can be expressed in multiple ways, we perform query expansion using the Unified Medical Language System (UMLS) to detect synonymous criteria. This is accomplished by expanding each criterion to include the set of all atoms in UMLS which have the same concept unique identifier (CUI) as the criteria. For example, “cerebrovascular disease” would be associated with 110 expansions, including “cerebral aneurysm”, “vascular ischemia”, “brain stem hemorrhage”, etc.
C. Relevance Models
The inclusion and exclusion criteria discerned from the query analysis were used by MERCuRY’s relevance models to assess the relevance of each EEG report against the given query. Two relevance models were considered (as illustrated in Figure 1): Case 1, which ignored the EEG signal fingerprints and Case 2, which incorporates them.
Case 1. This relevance model assigns a score to an individual EEG report based on the BM25F ranking function28. BM25F measures the relevance of an EEG report based on the frequency of mentions of each inclusion criterion and the absence of each exclusion criterion. Moreover, BM25F is capable of adjusting the score for each criterion based on the tiers in the posting list: that is, a criterion mention is scored according to both the polarity and the section in the document. Formally, for an EEG report r and a query q = {c1, c2, …} composed of individual inclusion and exclusion criteria (c), the BM25F relevance score is computed as:
where idf(c) is the inverse-document frequency of criterion c (i.e. the inverse of the number of documents mentioning c), K1 is a structuring parameter (in our case set to the standard28 value K1 = 1.2) and is a tier-normalizing criterion frequency measure. The tier-normalizing criterion frequency measure, , adjusts the frequency of criterion c in report r according to the polarity and section of each mention. Before defining this measure, we must account for the fact that query analysis described above considers two ways of representing inclusion and exclusion criteria – (a) by phrases and (b) by typed medical concepts; thus, the tier-normalizing criterion frequency measure changes depending on which of these methods is used:
(Case 1a) When an inclusion or exclusion criterion are expressed as a phrase, we defined (used by the BM25F function) in Equation (a), where xr,c,s,p is the number of occurrences of criterion c with polarity p in section s of report r; b is a normalizing parameter (in our case using the standard28 value b = 0.75), lr,p,s is the number of terms with polarity p in section s of report r, and ls,p is the average number of terms with polarity p in section s across all reports.
(Case 1b) In this case, each criterion is represented as a medical concept. For this method, the tier-normalizing criterion frequency measure is restricted only to the sections pertinent to the type of medical concept. That is, medical problems, and medical tests are only searched in the HISTORY and CORRELATION sections; medical treatments are searched in the MEDICATIONS and CORRELATION sections; while EEG activities and EEG events are searched only in the DESCRIPTION and IMPRESSION sections. Consequently, the tier-normalizing criterion frequency measure (used in the BM25F function) is computed using Equation (b) where t indicates a section pertinent to the type of the medical concept used to express the criterion c.
Case 2. The second relevance model considers both the information from EEG reports as well as the EEG signal fingerprints. It starts with the candidate patients discovered based on Case 1. The ranked list of patients is then updated based on the fingerprints associated with the most relevant patients’ EEGs. The rank updating procedure relies on two parameters: (1) γ, the rank threshold parameter indicating how many of the initially retrieved patients should be used for re-ranking (in our experiments we set γ = 5), and (2) δ, the fingerprint selection parameter which determines the number of similar fingerprints to consider for each patient (in our experiments we set δ = 3). The updated patient ranking is obtained as follows: for each patient px of the γ-highest ranked patients, we (i) find the fingerprint fx associated with px, (ii) use the hierarchy of EEG signal fingerprints (illustrated in Figure 2) from the multi-modal index to discover the σ most-similar fingerprints to fx, and (iii) insert the patients corresponding to these fingerprints into the ranked list of patients immediately after the patient px, thus generating a new ranked list of patients.
Evaluation
We evaluated two aspects of the MERCuRY system: (1) the overall quality of patient cohorts discovered by the system and (2) the quality of the polarity classifier used to process the EEG reports and to detect exclusion criteria in queries.
A. Evaluation of Patient Cohort Discovery
We primarily evaluated the MERCuRY system according to its ability to retrieve patient cohorts. To this end, we asked three neurologists to generate a set of 5 evaluation queries each and then used them for evaluation. A sample of these queries is illustrated in Table 1. For each query, we retrieved the ten most relevant patients as well as a random sample of ten additional patients retrieved between ranks eleven and one hundred. We asked six relevance assessors to judge whether each of these patients belonged or did not belong to the given cohort. Moreover, the order of the documents (and queries) were randomized and judges were not told the ranked position of each patient. Each query and patient pair was judged by at least two relevance assessors, obtaining an inter-annotator agreement of 80.1% (measured by Cohen’s kappa).
Table 1:
Example queries used to evaluate the MERCuRY system
| Patient Cohort Description (Queries) | |
|---|---|
| 1. | History of seizures and EEG with TIRDA without sharps, spikes, or electrographic seizures |
| 2. | History of Alzheimer dementia and normal EEG |
| 3. | Patients with altered mental status and EEG showing nonconvulsive status epilepticus (NCSE) |
| 4. | Patients under 18 years old with absence seizures |
| 5. | Patients over age 18 with history of developmental delay and EEG with electrographic seizures |
This experimental design allowed us to evaluate not only the set of patients retrieved for each cohort, but also the individual rank assigned to them. Specifically, we adopted standard measures for information retrieval effectiveness, where patients labeled as belonging to the cohort were considered relevant to the cohort query, and patients labelled as not belonging to the cohort were considered as non-relevant the cohort query. Because the relevance of a patient to a particular cohort can be difficult to automatically measure, we report multiple measures of retrieval quality.
Moreover, because our relevance assessments consider only a sample of the patients retrieved for each topic, we adopted two measures of ranked retrieval quality: the Mean Average Precision30 (MAP) and the Normalized Discounted Cumulative Gain31 (NDCG). The MAP provides a single measurement of the quality of patients retrieved at each rank for a particular topic. Likewise, the NDCG measures the gain in overall cohort quality obtained by including the patients retrieved at each rank. This gain is accumulated from the top-retrieved patient to the bottom-retrieved patient, with the gain of each patient discounted at lower ranks. Lastly, we computed the “Precision at 10” metric (P@10), which measures the ratio of patients retrieved in the first ranks which belong to the patient cohort. Although less statistically meaningful, the precision is the easiest to interpret in terms of clinical application in that a 100.00% Precision at 10 indicates that all of the patients returned above rank 10 completely satisfy all the criteria of the given cohort. By comparison, the other measures indicate the quality of the ranking produced by our system such that the MAP and NDCG scores capture the degree that a patient retrieved at each rank will more closely match the cohort criteria than patients retrieved at low ranks.
We measured the performance the MERCuRY system configured for the two relevance models illustrated in Figure 1: Case 1, in which only the EEG reports are considered and Case 2, in which both the EEG reports and EEG signal information is considered. In both cases, we considered both methods of representing inclusion and exclusion criteria: (a) using phrases composed of terms, and (b) using typed medical concepts. We compared these four combinations against three competitive baseline systems for text retrieval: Okapi BM2532 (BM25), language model retrieval using Dirichlet smoothing33 (LMD), and the Divergence from Randomness34 (DFR) framework using Poisson smoothing, Bernoulli and Zipfian normalization. Table 2 illustrates these results.
Table 2:
Quality of patient cohorts
| Relevance Model | MAP | NDCG | P @ 10 |
|---|---|---|---|
| Baseline 1: BM25 | 52.05% | 66.41% | 80.00% |
| Baseline 2: LMD | 50.37% | 65.90% | 80.00% |
| Baseline 3: DFR | 46.22% | 59.35% | 70.00% |
| MERCuRY: Case 1 (a) | 58.59% | 72.14% | 90.00% |
| MERCuRY: Case 1 (b) | 57.95% | 70.34% | 90.00% |
| MERCuRY: Case 2 (a) | 70.43% | 84.62% | 100.00% |
| MERCuRY: Case 2 (b) | 69.87% | 83.21% | 100.00% |
As shown, both configurations yield promising performance. Moreover, case 2 obtains the highest quality patient cohorts as measured by all three metrics. This shows that the multi-modal capabilities enabled by the EEG fingerprinting approach are able to identify patients who were missed when only the EEG reports were considered. The poor performance obtained by the baseline systems highlights the difficulty of automatically discovering patient cohorts. Moreover, the increase in performance obtained by MERCuRY Model 1 compared to Baseline 1 highlights the importance of medical language processing on EEG reports – particularly the role of the tiered index and the incorporation of exclusion spans. The highest performance was obtained by MERCuRY Model 2, showing the promise of including EEG signal information when discovering patient cohorts. This suggests that the content of EEG reports alone is not enough to adequately determine if a patient satisfies particular inclusion criteria. This finding is not surprising, as EEG reports were not written to completely replace the EEG signal, but rather to describe the important characteristics of the EEG recording which may be of interest to other neurologists. As such, EEG reports typically document only notable findings making it difficult to exclude patients based only on the text in the EEG reports. The superior performance obtained by MERCuRY Model 2 indicates that EEG fingerprinting is able to supplement the information in the EEG reports and bridge the gap between the high level description of EEG information in the text, and the low-level electrode potentials recorded in the EEG signal.
B. Evaluation of Polarity Classification
We evaluated the quality of our automatic polarity detection approach by performing 10-fold cross validation on the 2,349 manual annotations we produced and measured precision, recall and F1-measure, as shown in Table 3. We compared our classifier against two baseline classifiers, (a) “Baseline: Word Only” which uses only word features, and (b) “Baseline: UMLS Only” which uses only UMLS concept features. The MERCuRY classifier obtains substantially higher performance. Moreover, the poor performance of the baseline systems suggests that determining exclusion spans in text requires more information that lexical context and can be improved by incorporating NegEx and medical ontologies.
Table 3:
Polarity classification performance
| Label | Precision | Recall | F1-Measure |
|---|---|---|---|
| Baseline: Word Only | 24.50 | 29.65 | 59.35 |
| Baseline: UMLS Only | 37.08 | 14.22 | 20.55 |
| MERCuRY | 86.82 | 70.10 | 76.20 |
Discussion
In terms of polarity classification, the most common types of error were due to confusion regarding the exact boundary of negative regions of text, for example, the sentence “No focal or epileptiform features were identified in this record” was classified such that only “no focal” was negative, rather than the entire phrase “no focal or epileptiform features.” This indicates a failure by the incorporated standard natural language processing modules (part of speech and phrase chunking) to adapt to the clinical domain. One obvious path to improvement would be to annotate basic linguistic information on clinical documents – particularly on EEG reports.
Another common type of error was related to the binary granularity of polarity. For example, the excerpt there is a suggestion of a generalized spike and wave discharge in association with photic stimulation” the phrase “generalized spike and wave discharge” was labelled as having a negative polarity, despite the physician clearly indicating the possibility of such an activity. This implies that future work would be well supported by a more fine-grained approach to capturing the physicians’ beliefs, for example, by considering the assertions used in the 2010 i2b2 challenge10. Unfortunately, introducing assertions requires overcoming additional barriers including increased risk of misclassification, and accounting for the degree of similarity between different assertion values.
In terms of patient cohort retrieval, it is clear that the two methods of representing inclusion and exclusion criteria – (a) using phrases of terms and (b) using typed concepts – do not provide any significant changes to cohort performance. Based on our analysis, we believe this is primarily due to the fact that there is little ambiguity in the types of concepts used in EEG reports: a particular phrase or term (e.g. heart attack) was always associated with the same concept type. Moreover, the types of concepts are almost completely restricted by the section they occur in (e.g. EEG activities and events do not occur in the HISTORY, MEDICATION, or CORRELATION sections). This suggests that considering EEG concepts alone does not provide any additional value to considering terms directly. Moreover, because the index records positional information, multi-term concepts (e.g. “slow and sharp wave”) are handled identically to multi-term phrases. Despite this, a number of errors were observed. First, neither phrase chunking nor concept detection is sufficient to fully capture the semantics of all inclusion criteria. For example, epileptiform activities were often described as attributes of a particular wave (e.g. “slow rhythmic delta [waves]”) where the individual concept (i.e. “delta [waves]”) is far less meaningful than its attributes (“slow” and “rhythmic”). This suggests that performance can be improved by not only accounting for the attributes of epileptiform activities but by adjusting the relevance model to ensure that mentions of attributes actually modify the correct term - that is, to ensure that “slow” actually modifies the same wave as “rhythmic”.
Finally, the substantial increase in performance when using the full multi-modal index shows that EEG fingerprints are able to recover relevant information omitted from EEG reports. Unfortunately, as the rank of retrieved patient’s decreases, the quality of the cohort obtained by finding similar patients using EEG fingerprints decreases. We investigated multiple values of λ (the number of patients to be used for re-ranking) and σ (the number of similar fingerprints to retrieve), and found that increasing these values can result in a decrease in performance. Regardless, we did observe that the fingerprints often identify patients which were not retrieved using the report text alone. Moreover, we believe that by further refining the fingerprints we can improve the quality of patients retrieved with higher values of λ.
Conclusion
In this paper we describe a patient cohort retrieval system that relies on a multi-modal multi-tiered index that organizes clinical information automatically processed from a big data resource of EEGs. Generating the index involved both medical language processing on EEG reports, but also a novel and highly efficient representation of the EEG signal recordings provided by a highly-performant Long Short Term Memory network. When evaluating the quality of patient cohorts obtained when considering both EEG reports and signal recordings, we have a Mean Average Precision of 70.43%. This high performance highlights the promise of multi-modal retrieval from text and signal data. The remaining barriers of high-accuracy patient cohort identification from EEGs that need to be removed will rely on: (1) incorporating a more fine-grained representation of inclusion and exclusion semantics discerned from EEG reports, (2) extending medical language processing for capturing spatial and temporal information, and (3) tightly correlating the information from EEG reports with the EEG signal recordings. In future work, we plan to address these barriers using recent developments in neural learning.
Acknowledgements
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under award number 1U01HG008468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Smith SJM. EEG in the diagnosis, classification, and management of patients with epilepsy. J Neurol Neurosurg Psychiatry. 2005 Jun.76(2):ii2–7. doi: 10.1136/jnnp.2005.069245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beniczky S, Hirsch LJ, Kaplan PW, Pressler R, Bauer G, Aurlien H, et al. Unified EEG terminology and criteria for nonconvulsive status epilepticus. Epilepsia. 2013 Sep.54(6):28–9. doi: 10.1111/epi.12270. [DOI] [PubMed] [Google Scholar]
- 3.Edinger T, Cohen AM, Bedrick S, Ambert K, Hersh W. Barriers to retrieving patient information from electronic health record data: failure analysis from the TREC medical records track. AMIA; 2012. p. 180. [PMC free article] [PubMed] [Google Scholar]
- 4.England MJ, Liverman CT, Schultz AM, Strawbridge LM. A summary of the Institute of Medicine report. 2. Vol. 25. Epilepsy Behav; 2012. Epilepsy across the spectrum: Promoting health and understanding; pp. 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. JAMIA. 2014;21(2):221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voorhees EM, Hersh WR. Overview of the TREC 2012 Medical Records Track. TREC [Internet] 2012.
- 7.Voorhees EM. The trec medical records track. ACM-BCB; ACM; 2013. p. 239. [Google Scholar]
- 8.Demner-Fushmann D, Antani S, Simpson M, Thoma GR. pub2012019.pdf. J Comput Sci Eng. 2012 [Google Scholar]
- 9.Syeda-Mahmood T, Wang F, Beymer D, Amir A, Richmond N, Hashmi S. AALIM: Multimodal mining for cardiac decision support. IEEE. 2007:209–12. [cited 2016 Jul 4] [Google Scholar]
- 10.Uzuner Ö, South BR, Shen S, DuVall SL. i2b2/VA challenge on concepts, assertions, and relations in clinical text. JAMIA. 2011. 2010;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui L, Lhatoo SD, Zhang G-Q, Sahoo SS, Bozorgi A. EpiDEA: extracting structured epilepsy and seizure information from patient discharge summaries for cohort identification; AMIA; 2012. [PMC free article] [PubMed] [Google Scholar]
- 12.Sahoo SS, Lhatoo SD, Gupta DK, Cui L, Zhao M, Jayapandian C, et al. Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care; JAMIA; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harati A, Choi S-M, Tabrizi M, Obeid I, Picone J, Jacobson MP. The Temple University Hospital EEG Corpus. GlobalSIP. 2013; IEEE.2013. [Google Scholar]
- 14.Anonymous. Guideline 7: Guidelines for writing EEG reports. Am Electroencephalogr Soc. 2006;23(2):118. doi: 10.1097/00004691-200604000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan PW, Benbadis SR. How to write an EEG report: dos and don’ts; Neurology; 2013. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemp B, Olivan J. European data format “plus”(EDF+), an EDF alike standard format for the exchange of physiological data. Clin Neurophysiol. 2003;114(9):1755–1761. doi: 10.1016/s1388-2457(03)00123-8. [DOI] [PubMed] [Google Scholar]
- 17.Manning CD, Surdeanu M, Bauer J, Finkel JR, Bethard S, McClosky D. The Stanford CoreNLP Natural Language Processing Toolkit; ACL (System Demonstrations) [Internet]; 2014. pp. 55–60. [Google Scholar]
- 18.Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data; Proc 18th Int Conf Mach Learn ICML-2001; 2001. [Google Scholar]
- 19.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 20.Stone PJ, Bales RF, Namenwirth JZ, Ogilvie DM. The general inquirer: A computer system for content analysis and retrieval based on the sentence as a unit of information. Behav Sci. 1962;7(4):484–498. [Google Scholar]
- 21.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System; Methods Inf Med; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; Proc AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts K, Harabagiu SM. A flexible framework for deriving assertions from electronic medical records. JAMIA. 2011;18(5):568–573. doi: 10.1136/amiajnl-2011-000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes C, Vapnik V. Support vector machine. Mach Learn. 1995;20(3):273–297. [Google Scholar]
- 25.Kosko B. Bidirectional associative memories. Syst Man Cybern IEEE Trans On. 1988;18(1):49–60. [Google Scholar]
- 26.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 27.Muja M, Lowe DG. Fast Approximate Nearest Neighbors with Automatic Algorithm Configuration; VISAPP 1; 2009. [Google Scholar]
- 28.Zaragoza H, Craswell N, Taylor MJ, Saria S, Robertson SE. TREC. Citeseer; 2004. Microsoft cambridge at TREC 13: web and hard tracks; pp. 1–1. [Google Scholar]
- 30.Manning CD, Raghavan P, Schütze H. Vol. 1. Cambridge University Press; 2008. Introduction to information retrieval. [Google Scholar]
- 31.Järvelin K, Kekäläinen J. Cumulated gain-based evaluation of IR techniques. ACM Trans Inf Syst TOIS. 2002 [Google Scholar]
- 32.Robertson SE, Walker S, Jones S, Hancock-Beaulieu MM, Gatford M. Okapi at TREC-3; TREC; 1995. [Google Scholar]
- 33.Zhai C, Lafferty J. A study of smoothing methods for language models applied to ad hoc information retrieval. SIGIR; ACM; 2001. pp. 334–342. [Google Scholar]
- 34.Amati G, Van Rijsbergen CJ. Probabilistic models of information retrieval based on measuring the divergence from randomness. ACM Trans Inf Syst TOIS. 2002;20(4):357–389. [Google Scholar]