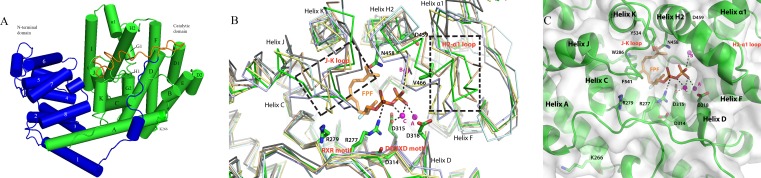
Figure 5. The homology model of PmSTS shows the structure domain and active site of the enzyme.
(A) The enzyme is made up of α-helices architecture structure to contain terpene synthase family N-terminal domain (blue) and C-terminal metal-binding domain (green). The truncated N-terminal (24 amino acid residues) disordered region of PmSTS_Δ24 is colored in orange. (B) Superimpose of PmSTS (green) to monoterpene synthase S. officinalis (+)-bornyl diphosphate synthase (SoBDS) (PDB ID:1N20 in purple) and M. spicata 4S-limonene synthase (MsLS) (PDB ID:2ONG in yellow), and sesquiterpene N. tabacum 5-epi-aristolochene synthase (NtEAS) (PDB:3M01 in brown), G. arboreum δ-cadinene synthase(GaDCS) (PDB ID: 3G4F in cyan) and A. annua α-bisabolol synthase (AaBOS) (PDB ID: 4FJQ in black), reveals the structural conserved RXR and DDXXD motifs, and flexible region (boxed) of J–K and H2-α1 loops at the active site. The ligand FPF and trinuclear metal cluster were adopted by superimposed NtEAS structure with PmSTS (C) The active site of PmSTS shows the ligand entrance pocket and the potential enzyme-substrate interactions. The three catalytic important Mg2+ ions are also shown in magenta sphere. The mutated residues K266E that found in L. lactis recombinant protein PmSTS K266E is as stick in helix A.