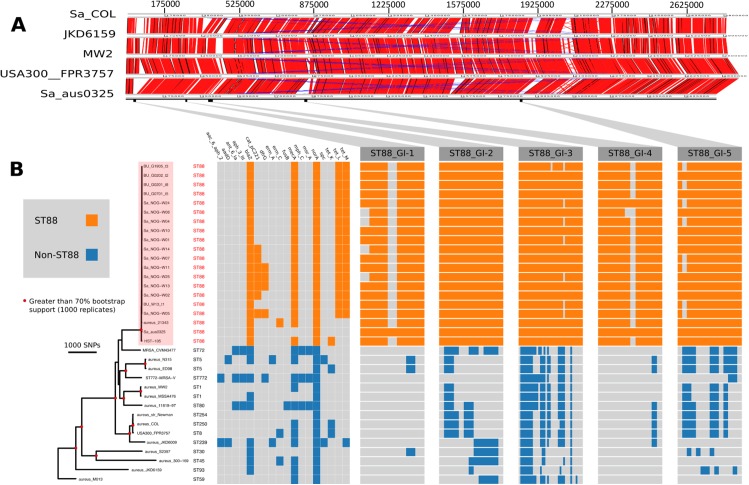
Figure 1. Comparative genomic analysis of S. aureus ST88.
(A) DNA–DNA comparisons visualized using the Artemis comparison tool of three CA-MRSA representative chromosomes and S. aureus COL against the complete chromosome of ST88 isolate AUS0325. (B) Core genome phylogeny and accessory genome elements identified among ST88 isolates. The phylogeny was based on an alignment of 71,862 non-recombinogenic core genome SNPs (indels excluded) and inferred using FastTree. Nodes with greater than 70% bootstrap support (1,000 replicates) are labelled with red dots. Antibiotic resistance genes were identified using Abricate (https://github.com/tseemann/abricate) and genomic islands (GIs) enriched among ST88 isolates were identified by ortholog comparisons using Roary and visualized using FriPan. CDS present in specific GI are listed in Table S1.