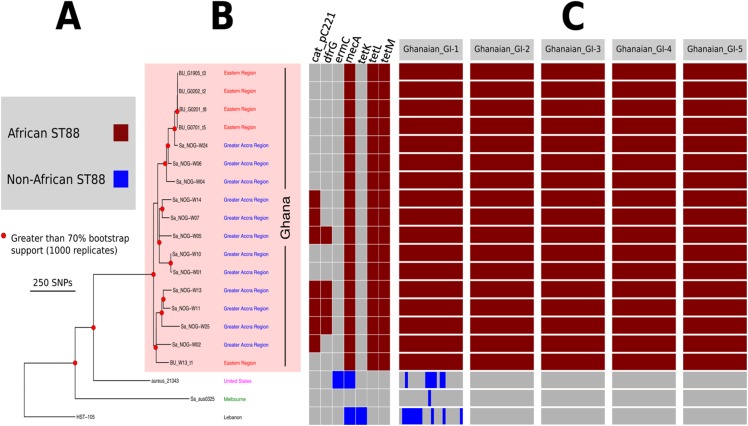
Figure 2. High resolution ST88 phylogeny and accessory genome analysis.
(A) Phylogeny inferred by read-mapping and variant identification among only ST88 genomes. Tree was produced using FastTree based on a pairwise alignment of 1,759 non-recombinogenic core genome SNPs among the 20 ST88 genomes. All major nodes in the tree (red circles) had greater than 70% bootstrap support (1,000 replicates). (B) Accessory gene content variation among the 20 ST88 genomes as assessed by ortholog comparisons using Roary. (C) Distinct genomic islands (GI) identified in Ghanaian isolates.