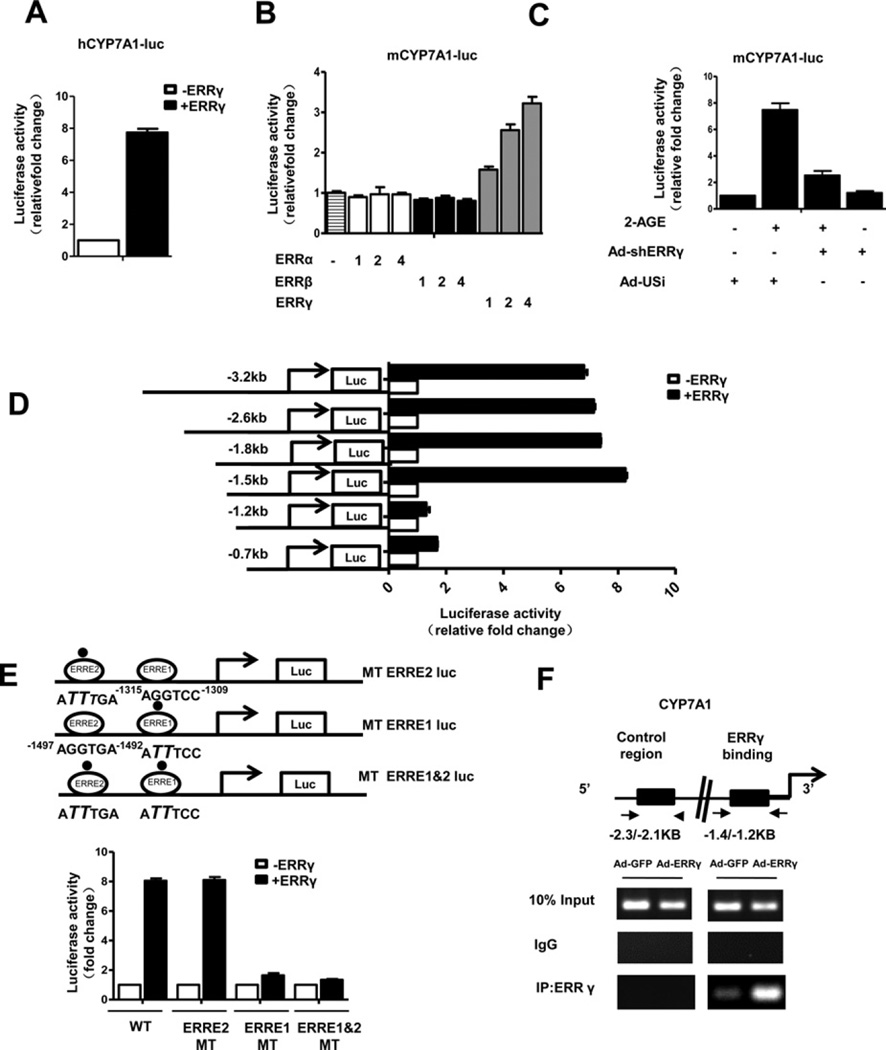
Figure 3. ERRγ activates the CYP7A1 gene promoter.
(A) 293T cells were transfected with hCYP7A1-luc (−1887 bp) and ERRγ expression vector. (B) 293T cells were transfected with mCYP7A1-luc (−3.2 kb to +234 bp) and different doses of the ERR subfamily cDNA expression vectors: 1:100 ng, 2:200 ng, 4:400 ng. (C) AML12 cells were infected with Ad-US or Ad-shERRγ for 48 h then transfected with mCYP7A1-luc (−3.2 kb to +234 bp) and treated with 2-AGE (10 µM). (D) 293T cells were transfected with deletion constructs of mCYP7A1-luc and ERRγ . (E) An ERRγ -binding site point mutation was made in mCYP7A1 (−1.5 kb)-luc as described in the ‘Materials and Methods’ section. 293T cells were transiently transfected with pCDNA3 – FLAG–ERRγ, mCYP7A1 (−1.5 kb to +234 bp)-luc (WT), mCYP7A1–mtERRE2-Luc, mCYP7A1–mtERRE1-Luc and mCYP7A1–mtERRE1 and 2-Luc. The cell lysates in (A–E) were utilized for luciferase and β-galactosidase assays. Experiments in (A–E) were conducted in triplicate and data are expressed as fold activation relative to the control. (F) ChIP assay. AML12 cells were infected with Ad-GFP or Ad-ERRγ for 48 h. Input represents 10% of purified DNA in each sample. Cell extracts were immunoprecipitated with anti-ERRγ antibody and purified DNA samples were employed for PCR with primers binding to ERRE1 (−1.4 kb to −1.2 kb) and distal site (−2.3 kb to −2.1 kb) on the CYP7A1 gene promoter.