Figure 2.
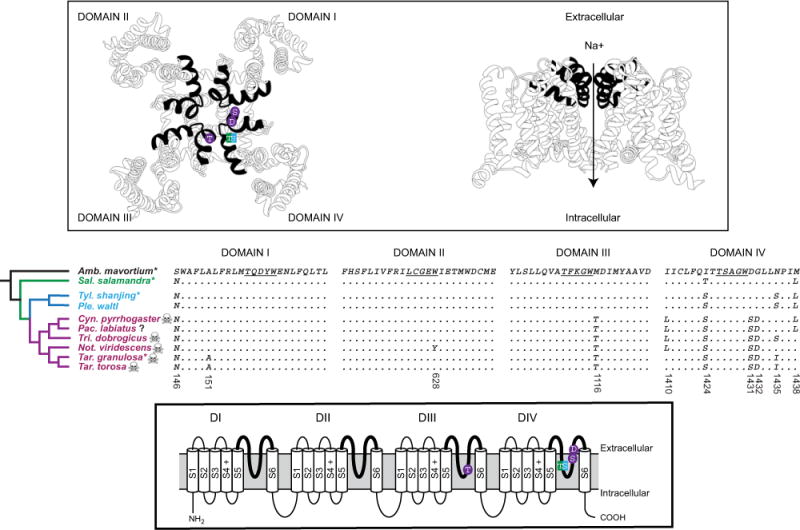
Substitutions in the salamander voltage-gated sodium channel, NaV 1.4. The phylogenetic distribution of amino acid substitutions in the outer pore of salamander NaV 1.4 (middle) is shown with a model of the TTX-binding site/outer pore (in black, above) and the transmembrane organization of NaV 1.4 (below). Candidate substitutions are also shown on the schematic structures of NaV 1.4 (above and below). Sequence alignments of the P1 and P2 helices with the selectivity filter linker (underlined) of all four domains are shown. Four S5–S6 extracellular linker regions (pore loops, bold lines) form the TTX-binding site as well as the part of the pore responsible for selective permeability to Na+ ions. Three substitutions (M116T, D1431S, and G1432D) are coupled with an extreme increase in the TTX resistance of modern newts, and Thr (true salamanders) and Ser (primitive and modern newts) substitutions at I1424 are associated with concomitant increases in TTX resistance of these lineages (Fig. 1). Substitutions and tree topology are color coded as in Fig. 1. Species for which TTX resistance was assayed (∗) and species known to possess TTX (skull) are indicated. The presence of TTX in Pachytriton is unknown (?). The upper model is based on the crystal structure of the bacterial sodium channel and Tikhonov and Zhorov (2012). Phylogentic relationships are inferred from Zhang et al. (2008) and are in agreement with the topology of salSCN4a (Fig. S1). Amino acid positions are based on sequence from Taricha torosa (GenBank numbers, TBD).
