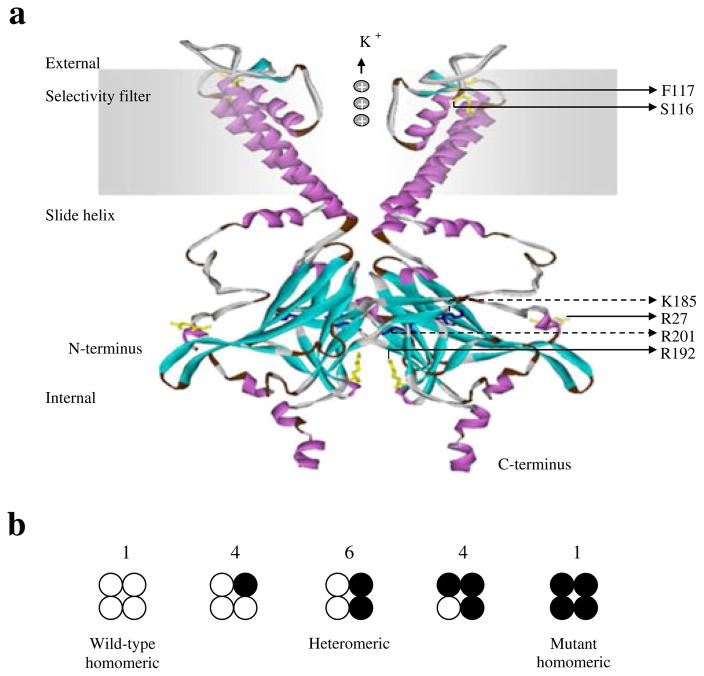
Fig. 2.
Final modelled ribbon structure of two subunits of Kir6.2, indicating the location of the novel KCNJ11 mutations. (a) The homology model of Kir6.2, which is quite similar to the previously reported structure of the other isoconformer, corresponds most closely to a truncated form of Kir6.2 expressed in the absence of SUR1. The labelled residues shown in yellow stick format are S116, F117, R27 and R192, which are indicated with solid arrows. The ATP-binding site is composed of the residues shown in the dark-blue stick model, R201 and K185, which are indicated with dashed arrows. (b) Schematic of the expected mixture of channels with different subunit compositions when wild-type and mutant Kir6.2 are coexpressed (as in the heterozygous state). The expected relative numbers of channel types for wild-type and mutant subunits are indicated above the figure, and segregate independently (i.e. follow a binomial distribution)