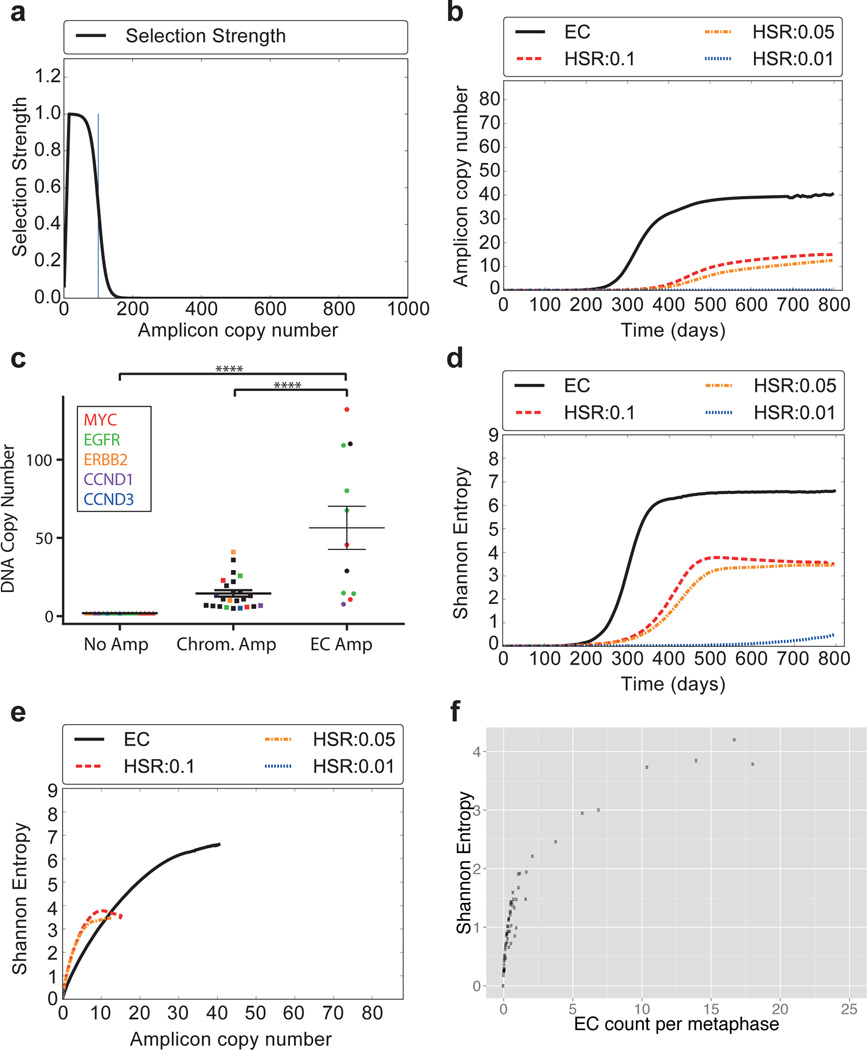
Fig. 4. Theoretical model for focal amplification via extrachromosomal (EC) and intrachromosomal (HSR) mechanisms.
Simulated change in copy number via random segregation (EC) or mitotic recombination (HSR), starting with 105 cells, 100 of which carry amplifications. a, The selection function f100(k) reaches maximum for k=15, then decays logistically. b, Growth in amplicon copy number over time. c, DNA copy number stratified by oncogene location. (p<0.001, ANOVA/Tukey’s multiple comparison). N=52; data points include top five amplified oncogenes, mean ± SEM. d, Change in heterogeneity (SI) over time. e, Correlation between copy number and heterogeneity. f, Experimental data showing correlation between ECDNA counts and heterogeneity matches the simulation in panel E.