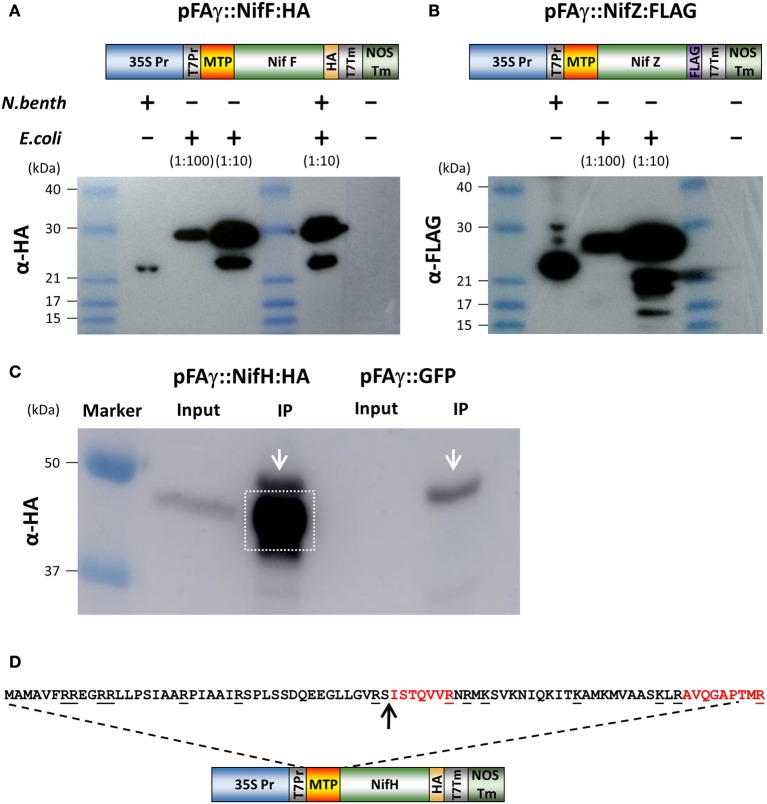
Figure 2.
Expression and processing of mitochondrially targeted Nif proteins in N. benthamiana. (A,B) Schematic of the pFAγ::NifF::HA and pFAγ::NifF::FLAG constructs used for N. benthamiana and E. coli expression, indicating the T7 promoter driving bacterial expression downstream of the 35S promoter for plant expression. Western blot analysis of pFAγ::NifF::HA or pFAγ::NifF::FLAG expression in N. benthamiana and E. coli. The + and − symbols above the lanes indicate the presence or absence, respectively, of N. benthamiana or E. coli protein extracts applied to the lanes; the extract dilution factors used for the bacterial extracts are indicated in brackets. (C) Image of Western blot for protein concentration of pFAγ::NifF::HA by anti-HA immunoprecipitation. Original protein extracts (Input) and IP eluate (IP) are shown. Background signal from the large-chain subunit of the HA antibody is marked with white arrow. The gel area excised for protein microsequencing is indicated by the white box. (D) Schematic of the construct used to transiently express pFAγ::NifH::HA in N. benthamiana. Underlined residues in the nucleotide sequence indicate sites of proteolytic cleavage (carboxyl side) by trypsin. The arrow indicates the point of cleavage by the mitochondrial processing peptidase (MPP). The peptides ISTQVVR and AVQGAPTMR were detected by mass spectrometry.