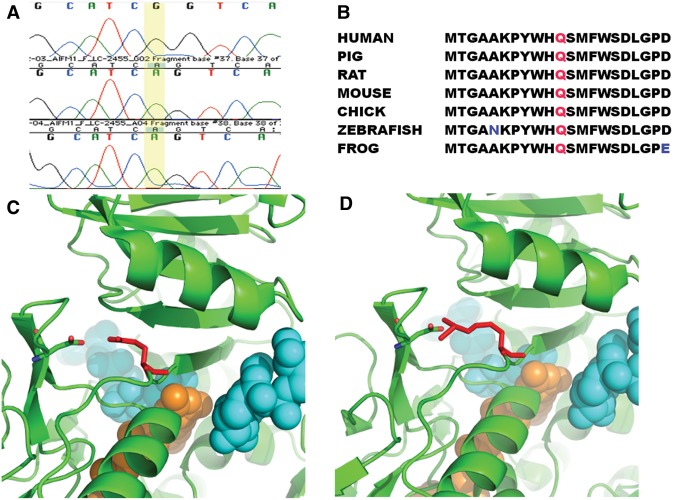
Figure 5.
Genetic variation in AIFM1 in affected patient. (A) Sanger sequencing results for affected region of AIFM1. The A→G mutation at position 1436 is present in the proband (top row) but not the parents (bottom two rows). (B) Alignment of affected amino acid across vertebrates with residue Gln479 colored in red. Nonconserved amino acids are in blue text. (C) Location of Gln479 variation. One AIFM1 binds to two NAD (colored in cyan) and one FAD (colored in orange). Residue Gln479 (colored in red) is near the binding of the FAD and one NAD. Its amino group interacts with the hydroxyl group of residue Asp443 electrostatically. (D) The NAD and FAD molecules are shown in Sphere. The mutation Gln479Arg does not change the charge status of this residue, but extends the length of the side chain of Gln479, which might disrupt the binding pocket of FAD and/or NAD. This change might affect the binding capability of AIFM1 to FAD and/or NAD.