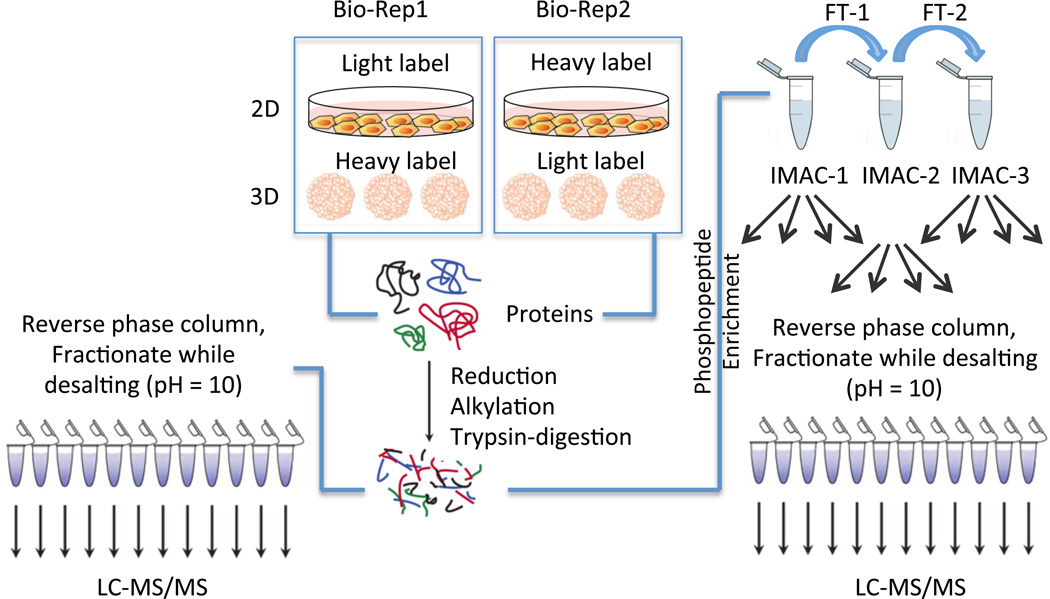
Figure 1.
Workflow for the experiments. HT29 cells were grown either as a 2D monolayer or in 3D spheroids in two biological replicates. In one biological replicate, the 3D spheroids were labeled with the heavy SILAC media while the monolayers were prepared in light SILAC media. Reverse labeling was used for the second biological replicate. Proteins were harvested from the 2D and 3D samples in each biological replicate and combined in a 1:1 ratio. The combined protein fractions were then aliquoted for proteomic and phosphoproteomic enrichment, with 500 µg used for the proteomic analysis and 3 mg of protein allocated for the phosphoproteomic enrichment. The proteins were then reduced and alkylated prior to tryptic digestion. For the proteomic analysis, peptides were fractionated on HLB reverse-phase columns and then analyzed by LC-MS/MS. For the phosphoproteomic analysis, peptides were enriched first by three rounds of IMAC, followed by high pH reverse phase fractionation prior to LC-MS/MS analysis, as described previously. (25)