Figure 4.
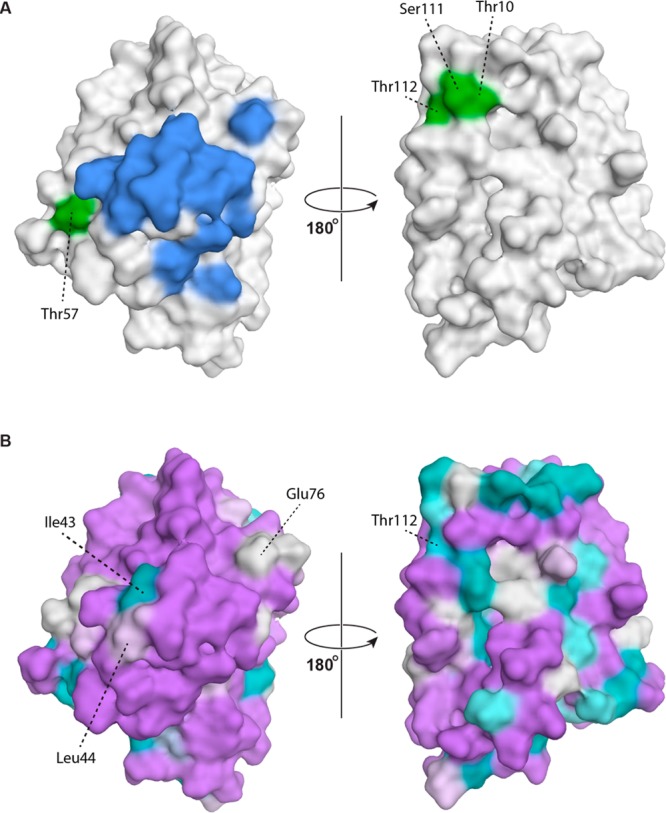
(A) Conformational epitopes of Mal d 1. Amino acid residues that correspond to the molecular interaction surface between monoclonal IgG BV16 and Bet v 1.0112 (residues Glu42–Thr52, Arg70, Asp72, Glu76, Ile86, and Lys97 in Mal d 1.0101) are colored in blue.45 Amino acid positions that were shown to be crucial for IgE recognition of Mal d 1 in mutational analyses (Thr10, Ile30, Thr57, Ser111, Thr112, and Ile113) are shown in green (Ile30 and Ile113 are located in the protein interior and do not contribute to the surface).13,34 (B) Amino acid similarities between Bet v 1.0101 and Mal d 1.0101 using a color gradient from lilac (highly similar) to teal (highly dissimilar). Epitope residues that are different between Bet v 1.0101 and Mal d 1.0101 are labeled. Similarities were calculated on the basis of substitution matrix scores (BLOSUM62) as implemented in MOE.32
