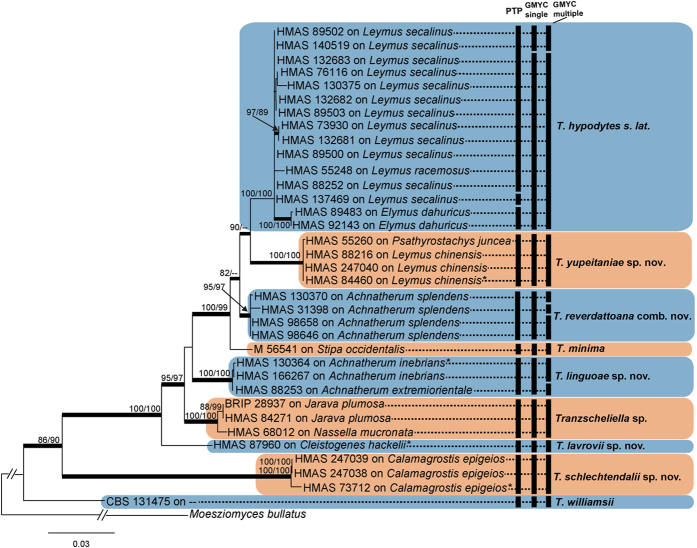
Figure 2. Phylogram obtained from a ML analysis based on the ITS and LSU sequence alignment.
Values above the branches represent ML bootstrap values (>75%) from RaxML and PhyML analyses respectively. Thickened branches represent Bayesian posterior probabilities (>0.95). The scale bar indicates 0.03 expected substitutions per site. Asterisk indicates type species. The first column depicts species recognized by PTP model. The second and third columns depict putative species recognized by the single-threshold and multiple-threshold GMYC model, respectively.