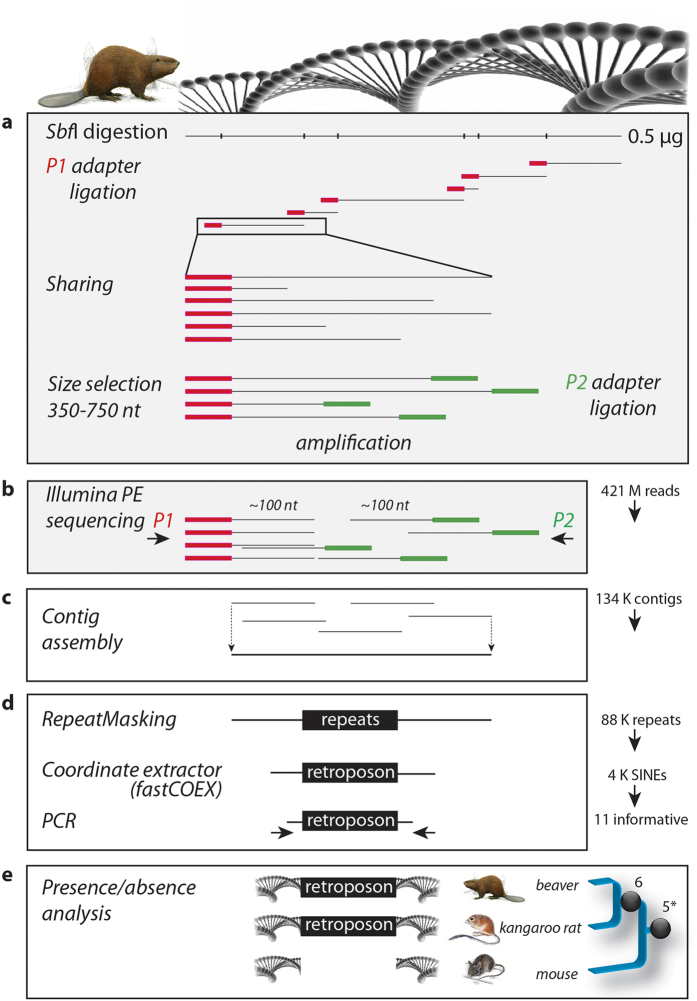
Figure 1. Snapshot of retroposon presence/absence analysis from Illumina paired-end reads (PE-RAD).
(a) Library construction: genomic SbfI digestion, P1 adapter ligation, random sharing (further shown for a selected single read), and size selection of 350–750 nt followed by the P2 adapter ligation and amplification steps. (b) Paired-end Illumina sequencing: the P1 and P2 sequencing primers generate shifted and partially overlapped ~100-nt reads. (c) The described PE-RAD sequences were taken from Senn et al.14 to build a gross genome assembly including ~134,000 fasta contigs >200 nt. (d) RepeatMasking revealed ~88,000 repeats and ~4,000 rodent-specific SINE loci. These loci were extracted from the fasta assembly using the coordinate extractor tool fastCOEX described in Materials and Methods. For control, the 11 phylogenetically informative loci were PCR-amplified/sequenced in beaver and comparatively sequenced in additional rodents. (e) Informative SINEs (black balls) are shared by beaver and kangaroo rat (6) or all species of the mouse-related clade (5 plus 1 additional previously published marker5 is denoted by *), indicating their close relationship. The in this study performed analyses are shown in white boxes. The rodent paintings were provided by Jón Baldur Hlíðberg.