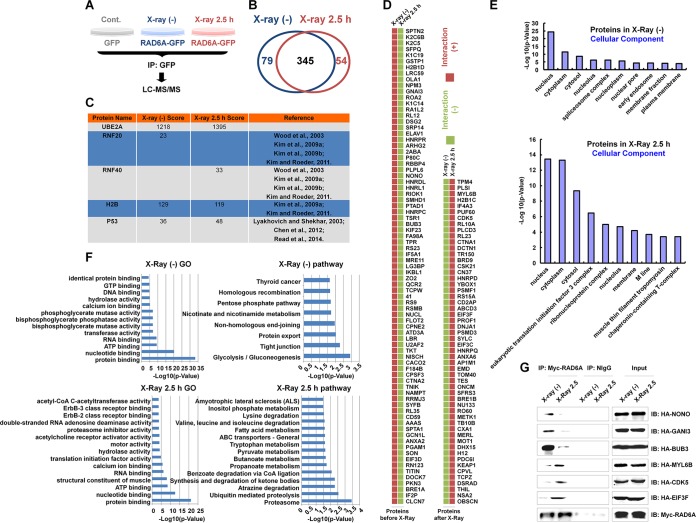
FIG 1.
Proteomic analysis of RAD6 interaction networks in the DNA damage response. (A) Schematic of the experimental procedures used to identify RAD6-interacting partners before and after DNA damage. HEK293T cells transfected with the GFP control or GFP-tagged RAD6A were treated with X-ray irradiation (X-ray 2.5 h) or not treated with X-ray irradiation [X-ray (−)], as indicated, at a dosage of 80 kV for 5 min, and the cells were recovered after 2.5 h. Cell extracts were prepared and subjected to co-IP assays with anti-GFP antibodies. The precipitated proteins were then subjected to mass spectrometry analysis. (B) After depletion of the nonspecific binding background by comparison with the GFP control, a number of RAD6A-interacting proteins were identified in cells without or with X-ray irradiation. The data are summarized in the Venn diagram. (C) The well-established RAD6-interacting proteins that were identified in our proteomics analysis are listed. The score refers to the obtained value analyzed by Mascot software on the basis of the original mass spectrum data. The following references mentioned in the figure appear at the indicated reference numbers in the References section: Wood et al., 2003, reference 13; Kim et al., 2009a, reference 9; Kim et al., 2009b, reference 14; Kim and Roeder, 2011, reference 15; Lyakhovich and Shekhar, 2003, reference 17; Chen et al., 2012, reference 16; and Read et al., 2014, reference 18. (D) All RAD6A-interacting proteins that appeared specifically under the control or X-ray irradiation conditions are listed. (E) The results of cellular component analysis of the RAD6A-interacting proteins that appeared under both control and X-ray irradiation conditions are shown. (F) The results of GO and pathway analyses of the RAD6A-interacting partners that appeared under both control and X-ray irradiation conditions are shown. (G) Some of the interacting proteins were selected and subjected to experimental validation. HEK293T cells were transfected with a Myc-tagged RAD6A construct combined with different randomly selected HA-tagged RAD6-interacting partners identified in our immunoprecipitation-mass spectrometry analysis for 48 h. Cells were then lysed and subjected to co-IP assays with anti-Myc antibodies. The normal mouse IgG (NIgG) antibodies were used as a negative control. The precipitated proteins were then subjected to Western blot analysis with the indicated anti-HA antibodies. CoA, coenzyme A; IP, immunoprecipitation; IB, immunoblotting.